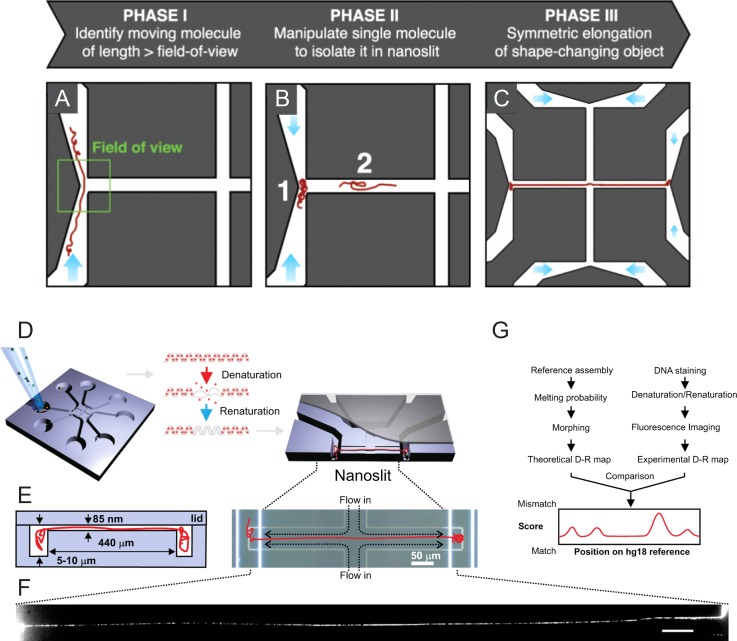
FIG. 10.
Fluidic chip design for DNA linearization and optical mapping. (a)–(c) Schematic representation of the main concept: (a) the DNA is pressure-driven through a microchannel to the nanoslit entrance, (b) loaded into the nanoslit by increasing the pressure inside the microchannel, and (c) elongated via pressure-driven flow from a second, perpendicular nanoslit. Fluorescence images from each phase can be used to automate the DNA manipulation procedure by real-time image processing. Reproduced with permission from Sørensen et al., Rev. Sci. Instrum. 86, 63702 (2015). Copyright 2015 AIP Publishing LLC.138 (d)–(f) The nanofluidic concept can be used for optical DNA mapping, as shown by the following example. (d) The chip is loaded with cell extract enriched in chromosomal DNA. Stained DNA is partially denatured and renatured in order to create a fluorescence pattern (see (f)). (e) The inlet ports of the chip connect to 5 - to 10 -μm-deep microchannels for DNA handling, which feed into an 85-nm shallow nanoslit. After stretching the DNA inside the nanoslit, the DNA is imaged. (f) The fluorescence image of a megabase pair-long DNA fragment shows a "barcode" where bright areas correspond to regions rich in cytosine-guanine, whereas dark areas correspond to regions rich in adenine-thymine. (g) The fluorescence pattern is compared to the pattern from the reference genome. Reproduced with permission from Marie et al., Proc. Natl. Acad. Sci., U.S.A. 110, 4893 (2013). Copyright 2013 National Academy of Sciences, USA.137