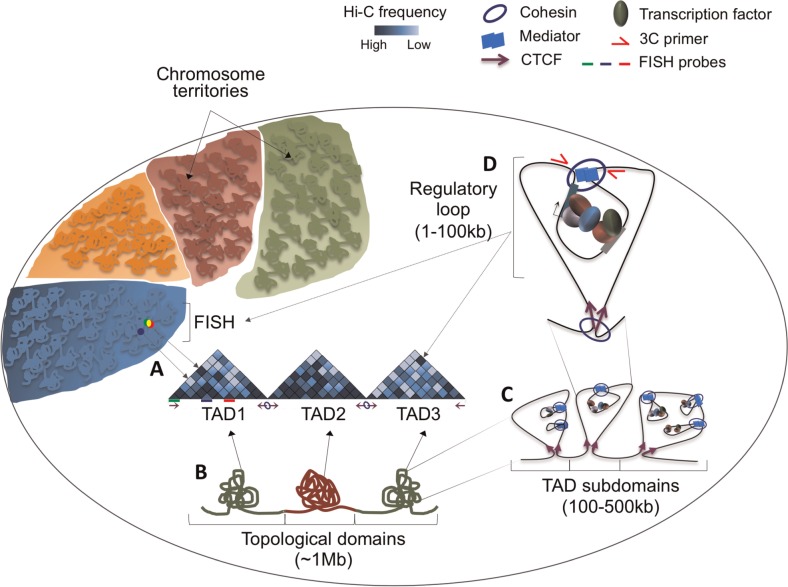
Figure 1.
Several layers of genome folding ensure higher order genome organization. Genomes are folded and distributed into CT. Within each CT, further folding delimited by converging CTCF binding sites (purple arrows) form ∼1 Mb topological domains (TADs). (A) FISH probes label three loci within TAD1. The close overlap between green and red FISH probes provides validation for a strong intra-TAD interaction between the genomic loci represented by green and red lines underneath the Hi-C heatmap. The high-frequency interaction is indicated as a dark blue bin in the heatmap. The blue FISH probe illustrates a locus that rarely interacts with the green locus within a folded chromosome, despite being in close linear proximity. (B) TADs associate with either open [26] or repressed [27] chromatin states. A specific locus intermingles more frequently within the same TAD, as represented in the Hi-C interaction heatmaps (darker blue: higher frequency of interactions; lighter blue: lower frequency of interaction). (C) Each TAD is further organized into subdomains that (D) are then folded in regulatory loops (dark blue circle in TAD3), where cis-regulatory regions are bound by transcription factors and contact promoters to regulate its function. Subdomains and regulatory loops are established and maintained by cohesin, mediator and converging CTCF. Interactions can be validated by FISH or by 3C. (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)