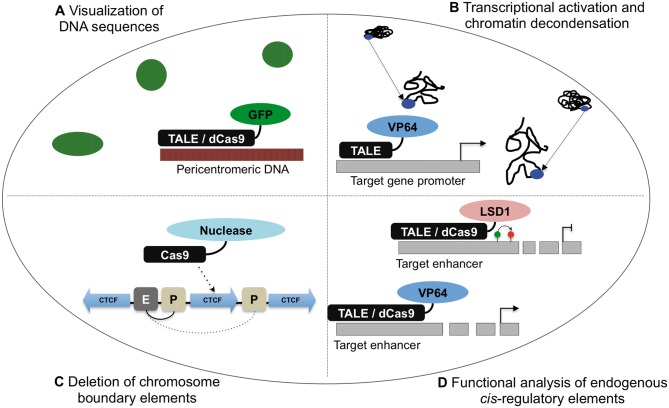
Figure 2.
Application of designer DNA-binding proteins to examine genome function. (A) TALE-GFP and dCas9-GFP fusion proteins can be targeted to DNA sequences, enabling the visualization of genome compartments in live cells. Example shown is for pericentromeric heterochromatin DNA, which cluster to form chromocentres in mouse cells. (B) The strong VP64 transactivator domain can be fused to TALE proteins, leading to transcriptional up-regulation of target genes. Example shown illustrates the repositioning within the nucleus of targeted gene regions and local chromatin decondensation that are caused by transcriptional up-regulation, as revealed by DNA FISH. (C) Targeting of Cas9-nuclease proteins to delete chromosome boundary elements. Example shown illustrates deletion of a CTCF binding site, which can result in the expansion of localized gene loops into neighbouring regions. E, enhancer; P, promoter. (D) TALE and Cas9 proteins can enable the investigation of cis-regulatory elements in several ways. Upper schematic represents TALE-LSD1 decommissioning of an enhancer element by demethylating the associated histone proteins, which can result in the transcriptional down-regulation of a nearby gene(s). Lower schematic shows targeting of TALE-VP64 or dCas9-VP64 to an enhancer element, which can result in the transcriptional up-regulation of a nearby gene(s). (A colour version of this figure is available online at: http://bfg.oxfordjournals.org)