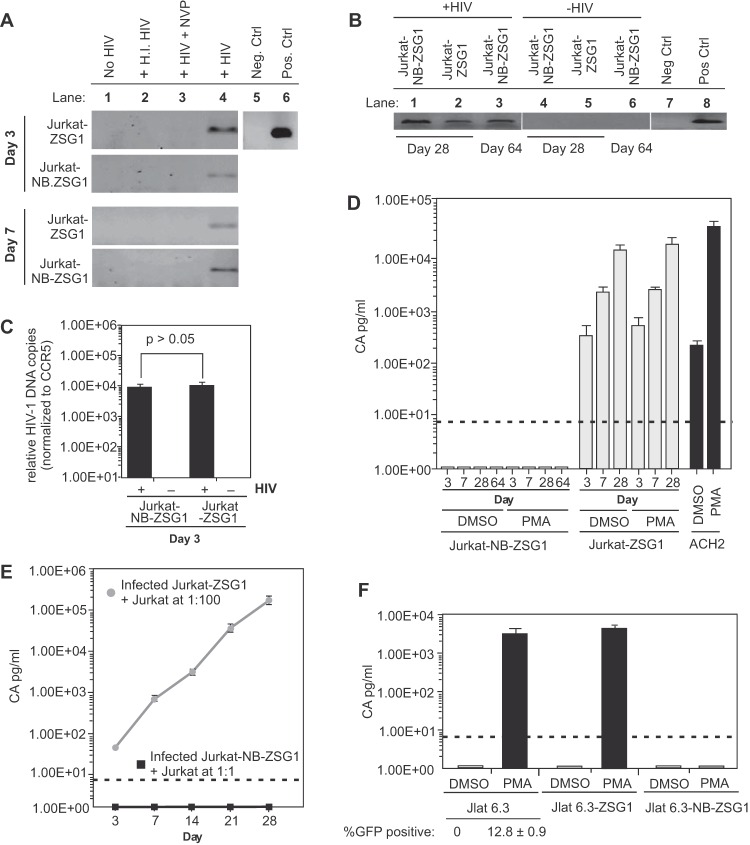
FIG 2 .
Jurkat-NB-ZSG1 cells harbor proviral HIV-1 DNA but no detectable viral mRNA. (A) PCR amplification of an HIV-1 env gene segment with total cellular DNA obtained from uninfected Jurkat, Jurkat-ZSG1, and Jurkat-NB-ZSG1 cells (lane 1) or from the same cell lines incubated with heat-inactivated (H.I.) virus (lane 2) or treated with or without nevirapine (NVP) for 2 h and infected with HIV-1NL4.3 (lanes 3 and 4). A PCR master mix alone was used as a negative (Neg.) control (lane 5) or with proviral DNA added as a positive (Pos.) control (Ctrl.) (lane 6). (B) Total cellular DNA obtained on days 28 and 64 from HIV-1-infected cell lines and assayed by endpoint PCR for env DNA. No viral DNA was detected in uninfected cells processed simultaneously (lanes 4 to 6). Negative and positive controls as in panel A are shown (lanes 7 and 8). (C) Total cellular DNA from day 3 HIV-1-infected or uninfected Jurkat-NB-ZSG1 and Jurkat-ZSG1 cells was assayed by Alu-PCR for integrated proviral DNA. The relative copy number was normalized to the CCR5 gene level in each sample. (D) On the days indicated, 1 nM PMA or DMSO (carrier) was added to Jurkat-NB-ZSG1 and Jurkat-ZSG1 cell cultures and incubated for 24 h. The soluble CA in the culture supernatant was then quantified by ELISA (the dotted line shows the limit of detection). ACH2 cell were used as a control for activation of HIV-1 gene expression by PMA. (E) Coculture of HIV-1-infected Jurkat-NB-ZSG1 or Jurkat-ZSG1 cells with uninfected Jurkat cells at a ratio of 1:1 or 1:100, respectively, for 28 days. The CA level in the supernatant was assayed by ELISA. (F) JLat 6.3, JLat 6.3-ZSG1, and JLat 6.3-NB-ZSG1 cells were incubated with 10 nM PMA for 24 h, and the CA levels in the supernatants were assayed by ELISA (the dotted line shows the limit of detection). The GFP-positive cell population was measured by flow cytometry. The assays in panels E and F were performed three times in triplicate, and the mean values and standard deviations are shown. Experiments representative of at least three independent experiments with similar results are shown. The P value in panel C was calculated with a Student t test.