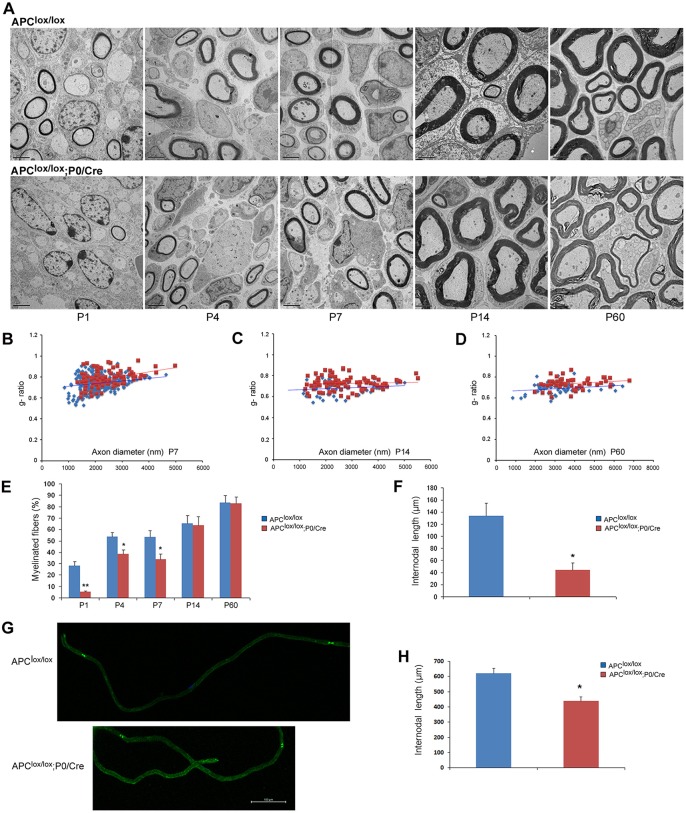
Fig. 2.
Loss of APC disrupts PNS myelination. The sciatic nerves of Apclox/lox;P0-Cre mutant and Apclox/lox control mice were analyzed by electron microscopy (EM). (A) The number of myelinated fibers is significantly reduced in the sciatic nerves of the Apclox/lox;P0-Cre mice at P1, P4 and P7 compared with control mice. Scale bars: 2 µm. (B-D) Apclox/lox;P0-Cre mice have a thinner myelin (higher g-ratio) compared with controls at P7 (B), P14 (C) and P60 (D). At P7, the mean g-ratio in the Apclox/lox;P0-Cre mice was 0.782 (±0.004) compared with 0.73 (±0.007) in the Apclox/lox mice, **P<0.001. At P14, the mean g-ratio in the Apclox/lox;P0-Cre mice was 0.72 (±0.006) compared to 0.68 (±0.009) in the Apclox/lox mice, **P<0.001. At P60, the mean g-ratio in the Apclox/lox;P0-Cre mice was 0.73 (±0.007) compared to 0.68 (±0.007) in the Apclox/lox mice, **P<0.001. (E) Apclox/lox;P0-Cre sciatic nerve contained fewer myelinated axons at P1, P4 and P7. *P<0.05, **P<0.001. (F) Three-dimensional EM images were acquired from sciatic nerve serial sections of Apclox/lox;P0-Cre and Apclox/lox (control) mice at P7, and individual nerve fibers were traced and analyzed in reconstructed EM images of both genotypes. The Apclox/lox;P0-Cre sciatic nerve contained much shorter internodes compared with controls. (G) Sciatic nerves of Apclox/lox;P0-Cre and Apclox/lox control mice at P60 were teased and immunolabeled for paranodin/Caspr, a marker of paranodal junctions to measure internodal length in adulthood. (H) Quantitative analysis showed that the Apclox/lox;P0-Cre sciatic nerves contained much shorter internodes compared with controls. For EM analysis, at least three mice were taken for each time point, per genotype. For g-ratio analysis, at least 50 axons from at least three mice per genotype were analyzed. The number of internodes analyzed in vivo in P7 animals was: Apclox/lox, n=9; Apclox/lox;P0-Cre, n=8; *P<0.05. The number of internodes analyzed in teased fibers of P60 animals was: Apclox/lox, n=46; Apclox/lox;P0-Cre, n=37; *P<0.05. Internodes from at least three mice per genotype were analyzed.