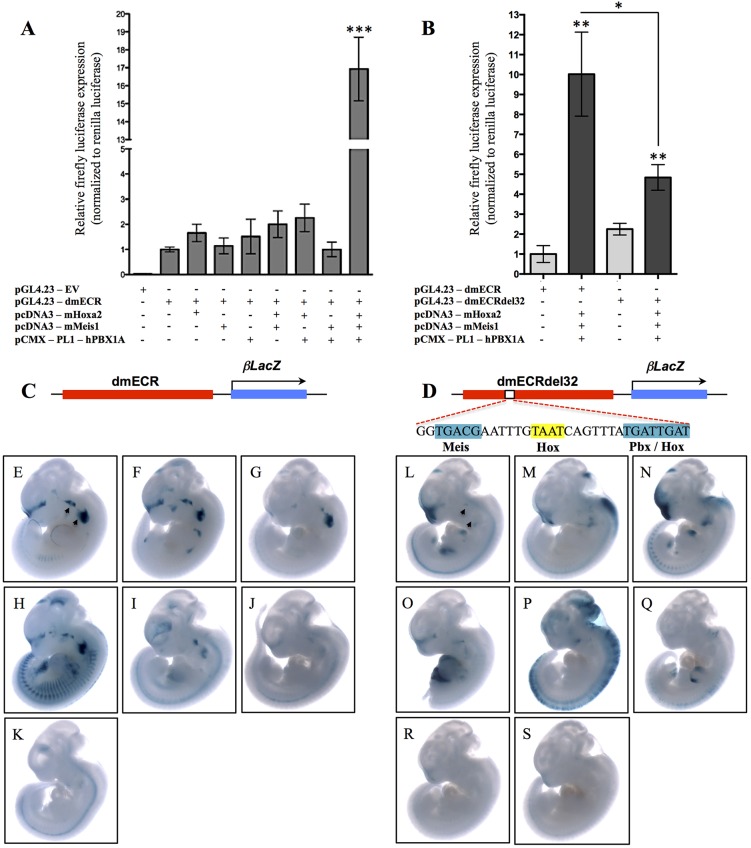
Fig. 8.
Hox-Pbx-Meis cooperatively bind to and regulate the dmECR via a 32 bp sequence. (A) Luciferase activity (relative firefly/Renilla levels) for the 594 bp dmECR sequence following transfection with the pcDNA3-mHoxa2, pcDNA3-mMeis1 or pCMX-PL1-hPBX1A expression vectors showed strong activation only in the presence of Hoxa2, Meis1 and PBX1A proteins (replicates n=3, Hoxa2 P=0.1386, Meis1 P=0.6913, PBX1A P=0.4998, Hoxa2+Meis1 P=0.1327, Hoxa2+PBX1A P=0.0862, Meis1+PBX1A P=0.9959, Hoxa2+Meis1+PBX1A ***P=0.0009). (B) Luciferase activity for the intact 594 bp dmECR sequence compared with the dmECRdel32 construct harboring the 32 bp deletion of the adjacent Hox-Pbx-Meis binding sites (see D). A significant reduction in enhancer activation from the dmECRdel32 construct compared with the intact dmECR construct was seen in the presence of Hoxa2, Meis1 and PBX1A proteins (replicates n=3, pGL4.23-dmECR+Hoxa2+Meis1+PBX1A P=0.0019, pGL4.23-dmECRdel32+Hoxa2+Meis1+PBX1A P=0.0031; pGL4.23-dmECR compared with pGL4.23-dmECRdel32 P=0.0152). (C-S) Transient transgenesis for the intact 594 bp dmECR construct (C) compared with the dmECRdel32 construct (D) showed almost complete loss of BA1/2 staining in E11.5 transient transgenic embryos generated from the dmECRdel32 construct (L-S, arrowheads) compared with the consistency of activity in these regions in transient transgenic embryos generated from the intact dmECR construct (E-K, arrowheads).