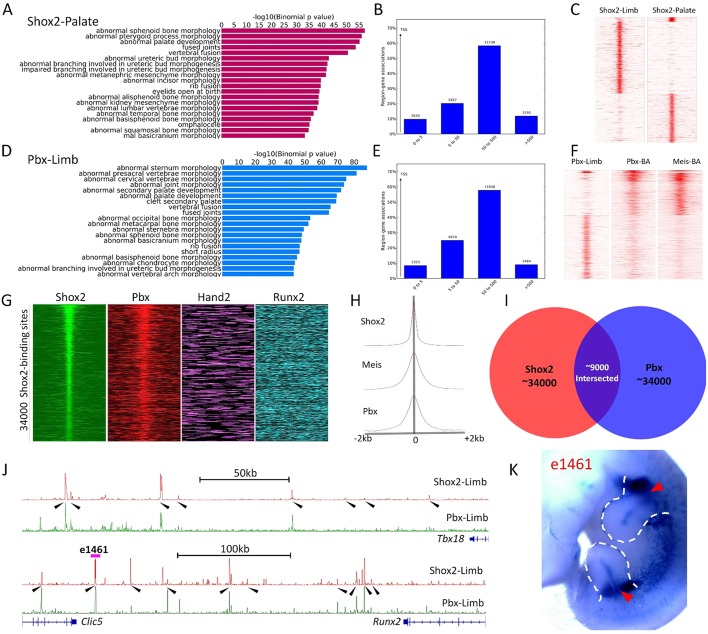
Fig. 6.
Genome-wide co-occupancy of Shox2 with Hox-TALE factors on osteogenic genes. (A-F), ChIP-Seq of Shox2 on the developing palate (A-C) and ChIP-Seq of Pbx on the developing limb (D-F) at E12.5. GO analysis and distance to TSSs histogram of Shox2-Palate (A,B) and Pbx-Limb (D,E) show similar functional and binding properties to Shox2-Limb ChIP-Seq data. K-mean cluster based on binding signal shows distinct binding preference of Shox2 occupancy in the limb compared with that in the palate (C). Similarly, Pbx preferentially binds to sites that are relatively weakly occupied by Pbx and Meis in the developing brachial arch (BA) (F). (G) Heat-map plot of binding signals of Shox2 (limb), Pbx (limb), Hand2 (limb) and Runx2 (osteogenic cell line) around Shox2 binding sites in E12.5 limb. (H) Aggregate plots on ChIP binding signal of Shox2 (limb), Pbx (limb) and Meis (BA) around Shox2 binding sites. (I) Venn diagram shows the ∼9000 co-binding sites of Shox2 and Pbx in proportion to the top ∼34,000 Shox2 and Pbx binding sites in E12.5 limb. (J) Shox2 and Pbx co-occupied enhancers cluster around Tbx18 and Runx2 (highlighted by arrowheads). (K) Representative transgenic embryo of e1461, a putative enhancer for Runx2 (indicated in J), shows proximal limb-specific activity (arrowheads).