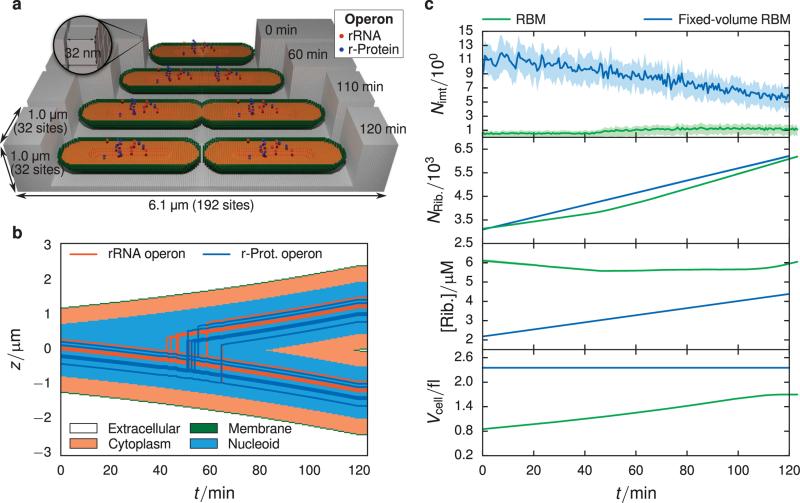
FIGURE 4.
(a) Schematic of geometry used in RBM simulations. The lattice is 32 × 32 × 192 sites in the x, y, and z directions respectively, with a lattice spacing of 32 nm. The simulation volume consists of 4 regions: (1) extracellular space (gray), (2) membrane (green), (3) cytoplasm (orange), and (4) nucleoid (not colored, found in center of cytoplasm). The initial length, 2.4 μm, and the width of the cell, 0.7 μm, were chosen from the previous experimental analysis (Section 2.1). The proportion of the nucleoid region to the cytoplasm is based on measurements of cryo-electron tomograms of slow-growing E. coli15. Operon species are placed within the nucleoid region based on their genomic loci and replicated at times computed from their genomic distance to the origin of replication. The position of the operon species is evolved in time such that the operons in the daughter cell are found in the same position as the operons in the mother cell. The cell volume grows constantly throughout the cell cycle at an exponential rate, where upon it divides into two daughter cells of length 2.4 μm. (b) Kymograph showing the evolution of spatial compartments and operon locations over one cell cycle. The jagged steps arise from the discreteness imposed by the 32 nm lattice. (c) Comparison between RBM (green) and fixed-volume RBM (blue) models using 16 replicates. Means are represented by solid lines and the interquartile range is given by the shaded area. There is an significantly lower average SSU intermediate count seen in the RBM compared to the fixed-volume RBM (top panel), which is a result of the changing cell volume. In the last three panels are plotted the absolute count of ribosomes (translating as well as dissociated), the absolute ribosome concentration, and the cell volume. The RBM produces ribosomes at approximately the same pace as volume expansion, leading to a constant ribosome concentration over the cell cycle.