Figure 2. Mettl3 is the active subunit, and Mettl14 catalytic motif is dispensable.
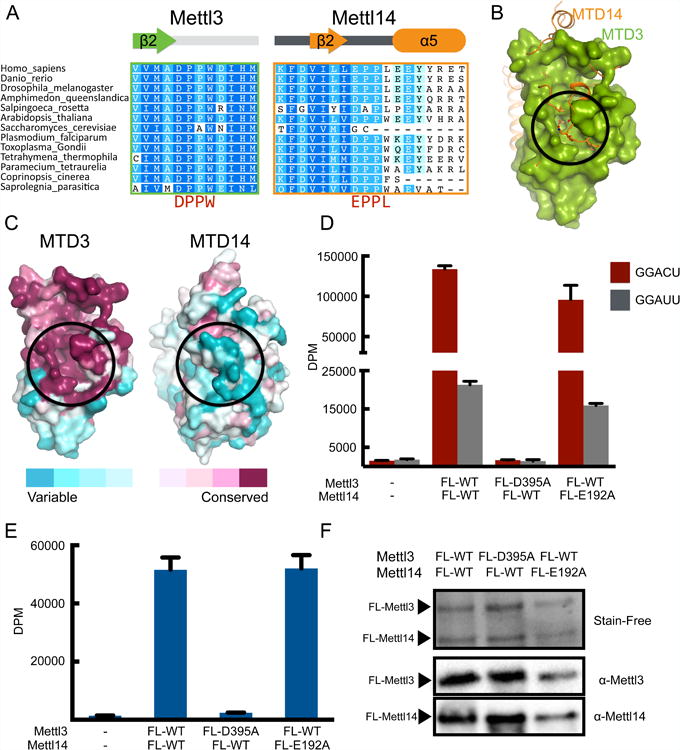
(A) Sequence alignment of the region containing the catalytic motifs DPPW (Mettl3) and EPPL (Mettl14). The secondary structures of the segments are indicated above the sequences.
(B) Superimposition of MTD3 (green surface representation) and MTD14 (orange cartoon and sticks representation). The proposed catalytic cavity (black circle) is more occluded in MTD14.
(C) Sequence conservation shown on surface representation of MTD3 and MTD14. Both MTD3 and MTD14 are colored according to the conservation score. The proposed active site is marked with a black circle.
(D) In vitro methyltransferase activity of the full-length Mettl3/Mettl14 complexes expressed in E. coli, with indicated point mutations for each polypeptide. Bars correspond to amounts of tritium incorporated into methylated RNA substrates shown as disintegrations per minute (DPM) with cognate (GGACU=red) or mutant (GGAUU=gray) sequence. Data shown as mean +/- SD from three replicates.
(E) In vitro methyltransferase activity of the full-length Mettl3/Mettl14 complex with indicated point mutations purified from HEK293 cells (blue bars). Data shown as mean +/- SD from three replicates.
(F) SDS-PAGE analysis (visualized by Stain-Free dye) followed by western blot of the proteins expressed in HEK293 cells used in the in vitro methylation assay in Figure 2E.
See also Figures S2 and S3.
