Figure 2.
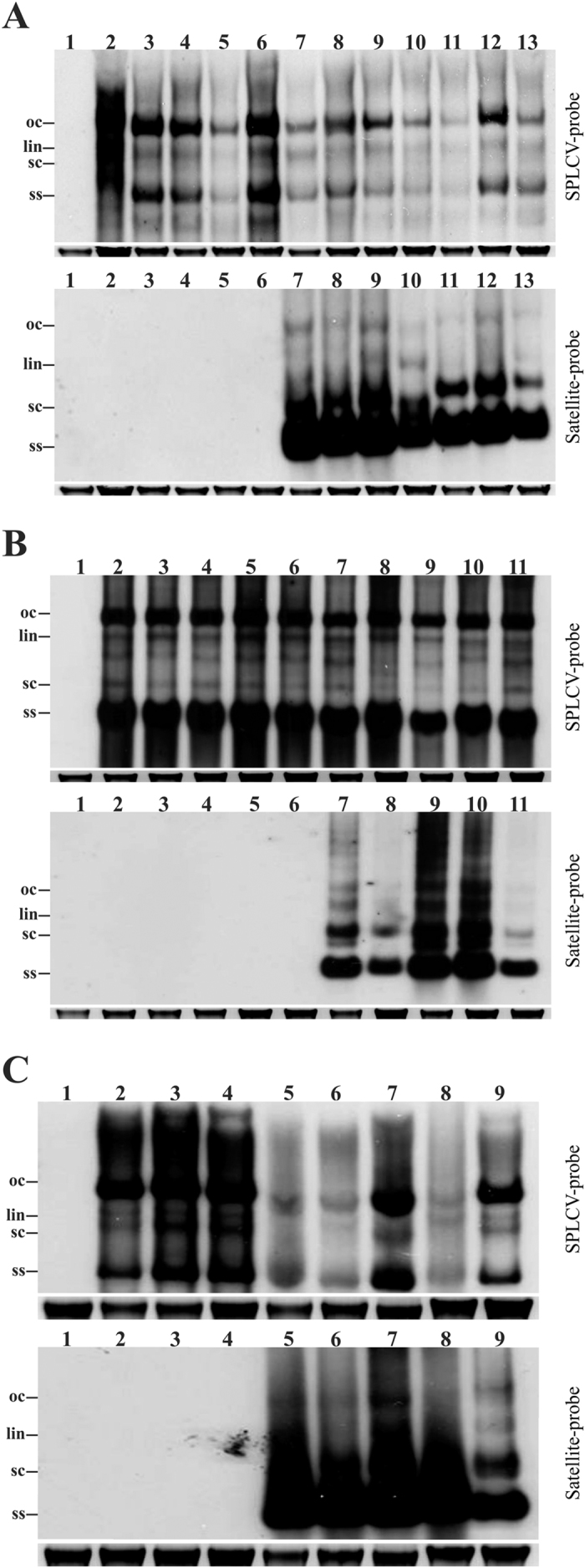
Southern blot analysis of DNA samples extracted from plants infected with SPLCV and satellites by either Agrobacterium-mediated inoculation of N. benthamiana (A) Agrobacterium-mediated inoculation of I. setosa (B) or insect transmission to I. setosa (C). Blots were probed for the presence of SPLCV (upper panel) and SBG51 (lower panel) in each case. (A) The DNA extracts were from a healthy non-inoculated N. benthamiana (lane 1) and N. benthamiana plants inoculated with SPLCV (lanes 2–6), SPLCV/DIM-SBG51 (lanes 7–10) and SPLCV/DIM-SBG59 (lanes 11–13). (B) The DNA extracts were from a healthy non-inoculated I. setosa (lane 1) and I. setosa plants inoculated with SPLCV (lanes 2–6) and SPLCV/DIM-SBG51 (lanes 7–11). (C) The DNA extracts were from a healthy non-inoculated I. setosa plant (lane 1) and I. setosa plants inoculated with B. tabaci insects fed on a SPLCV/SBG51 infected plant (lanes 2–9). The positions of viral single stranded (ss) super-coiled (sc), linear (lin) and open-circular (oc) DNAs are indicated. The ethidium bromide-stained genomic DNA band on the gel is shown below the Southern blot in each case to confirm equal loading. DNA was extracted at 20 dpi and 6 μg of total DNA were loaded in each case.
