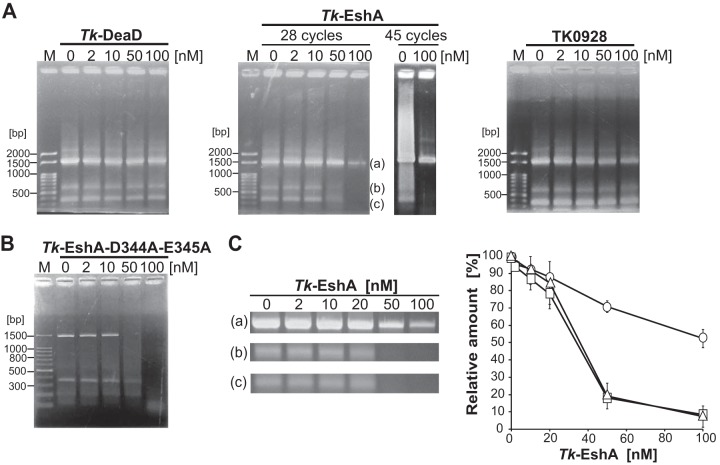
FIG 3.
Effect of thermostable helicases on PCR specificity. (A) A 1,498-bp region of 16S rRNA genes from T. kodakarensis was amplified by 28 or 45 repeating cycles in the presence of Tk-DeaD (left panel, 28 cycles), Tk-EshA (center panel, 28 or 45 repeating cycles), or TK0928 (right panel, 28 repeating cycles). The concentration of the helicases was 0, 2, 10, 50, or 100 nM. DNA size markers are shown in lane M. (B) A 1,498-bp region of 16S rRNA genes from T. kodakarensis was amplified by 28 repeating cycles in the presence of 0, 2, 10, 50, or 100 nM Tk-EshA-D344A-E345A. The 100-bp DNA ladder is shown in lane M. (C) Relative amounts of PCR products amplified by 28 repeating cycles upon addition of Tk-EshA. In the gels shown in the center of panel A, band a is the target product and bands b and c are nonspecific products. The relative amounts of desired and nonspecific amplification products were measured using ImageJ software. In the graph shown on the right, the circles, squares, and triangles denote the relative amounts of products a, b, and c, respectively. The relative amounts of PCR products were normalized based on the intensities of the bands. Each band (a, b, or c) was in the absence of Tk-EshA, which was set to 100%.