Abstract
Nuclear receptor (NR) signaling pathways impact cellular function in a broad variety of tissues in both normal physiology and disease states. The complex tissue-specific biology of these pathways is an enduring impediment to the development of clinical NR small-molecule modulators that combine therapeutically desirable effects in specific target tissues with suppression of off-target effects in other tissues. Supporting the important primary research in this area is a variety of web-based resources that assist researchers in gaining an appreciation of the molecular determinants of the pharmacology of a NR pathway in a given tissue. In this study, selected representative examples of these tools are reviewed, along with discussions on how current and future generations of tools might optimally adapt to the future of NR signaling research.
Introduction
The Nuclear Receptor Superfamily
The 48 proteins of the nuclear receptor (NR) superfamily function as ligand-dependent transcription factors for a diverse set of fat-soluble hormones, vitamins, and dietary lipids (Mangelsdorf et al., 1995). Included in this family are receptors for endocrine steroids (i.e., corticosteroids, progesterone, androgens, and estrogens), fat-soluble vitamins A and D, thyroid hormone, fatty acids, oxysterols, bile acids, and numerous environmental endocrine-disrupting chemicals and xenobiotics. Additional members of this family are referred to as orphan receptors because their ligands remain uncharacterized. As directly druggable regulators of gene expression, NRs and their transcriptional coregulators (Glass et al., 1997; McKenna et al., 1999) are pharmacologically prominent targets for the development of small-molecule therapeutics in a variety of inflammatory, neoplastic, and metabolic conditions (Glass and Ogawa, 2006; Schulman, 2010; Safe et al., 2014).
Biology of NR Signaling Pathways
Signaling pathways involving NRs, their cognate physiologic ligands, and coregulators coordinate the organ- and tissue-specific expression of genes across diverse physiologic systems. Processes regulated by NR signaling pathways include mammalian embryonic development [retinoic acid receptor and all-trans retinoic acid pathway (Mark et al., 2009)]; reproduction [estrogen, progesterone, and androgen receptor pathways (Carpenter and Korach, 2006; Zhao et al., 2008; Rubel et al., 2010)]; metabolism and the inflammatory response [glucocorticoid receptor and peroxisome proliferator-activated receptor (PPAR) subfamily pathways (Pyper et al., 2010; Giordano Attianese and Desvergne, 2015; Granner et al., 2015; Janani and Ranjitha Kumari, 2015)]; and the immune system and bone homeostasis [vitamin D receptor pathway (Christakos et al., 2016)]. Although a full discussion of the biology of NR coregulators is beyond the scope of this minireview, significant findings in this area are the roles of nuclear corepressors 1 and 2, and Mediator 1, and members of the steroid receptor coactivator family in embryonic development, the cardiovascular system, metabolism, and reproduction (McKenna et al., 1999; Giudici et al., 2016). Although NR signaling pathways are commonly named for their principal receptors, ligands and coregulators are key regulatory nodes, and the mechanism by which each pathway communicates the afferent physiologic signal varies between distinct tissues and cell types.
Clinical Pharmacology of NR Signaling Pathways
The extensive biologic footprint of NR signaling pathways is reflected in the intense interest they command as drug targets in a wide variety of human diseases and disorders. The clinical pharmacological agents that target NRs—popularly known as selective receptor modulators—selectively agonize or antagonize their cognate receptors in a tissue-, cell type–, and promoter-specific manner [comprehensively reviewed by Burris et al. (2013)]. Selective estrogen receptor modulators have found clinical application in estrogen receptor (ER)-positive [tamoxifen (Burris et al., 2013)] and metastatic [toremifene (Mustonen et al., 2014)] breast cancer, osteoporosis [raloxifene (Gizzo et al., 2013)], and vaginal atrophy [lasofoxifene (Pinkerton and Stanczyk, 2014)]. Given their robust antagonism of these signaling conduits in cells mediating the immune and inflammatory responses—B cells, T cells, and macrophages—a variety of glucocorticoid receptor-specific selective receptor modulators is in active clinical use for inflammatory and allergic conditions of the respiratory system (e.g., asthma, rhinitis) and skin (acne, psoriasis) and autoimmune disorders (rheumatoid arthritis), and to suppress local inflammatory responses in musculoskeletal injuries (Burris et al., 2013). The best-characterized—and most controversial—selective modulators of PPAR-γ are the thiazolidinediones, including rosiglitazone, pioglitazone, and troglitazone, which have been used as insulin-sensitizing hypoglycemic agents in the treatment of type 2 diabetes (Soccio et al., 2014). The undesirable side effects of selective estrogen receptor modulators, such as the increased risk of endometrial cancer associated with Tam use (Burris et al., 2013); incidents of heart failure, bone fracture, weight gain, and liver dysfunction associated with selective peroxisome proliferator reecptor modulators (Burris et al., 2013); and the effects of glucocorticoid receptor-specific selective receptor modulators on fluid retention, weight gain, and hypertension (Burris et al., 2013), are a signal reminder of the highly nuanced and contextual nature of NR signaling pathway pharmacology.
Research Resources for Analysis of NR Signaling Pathways
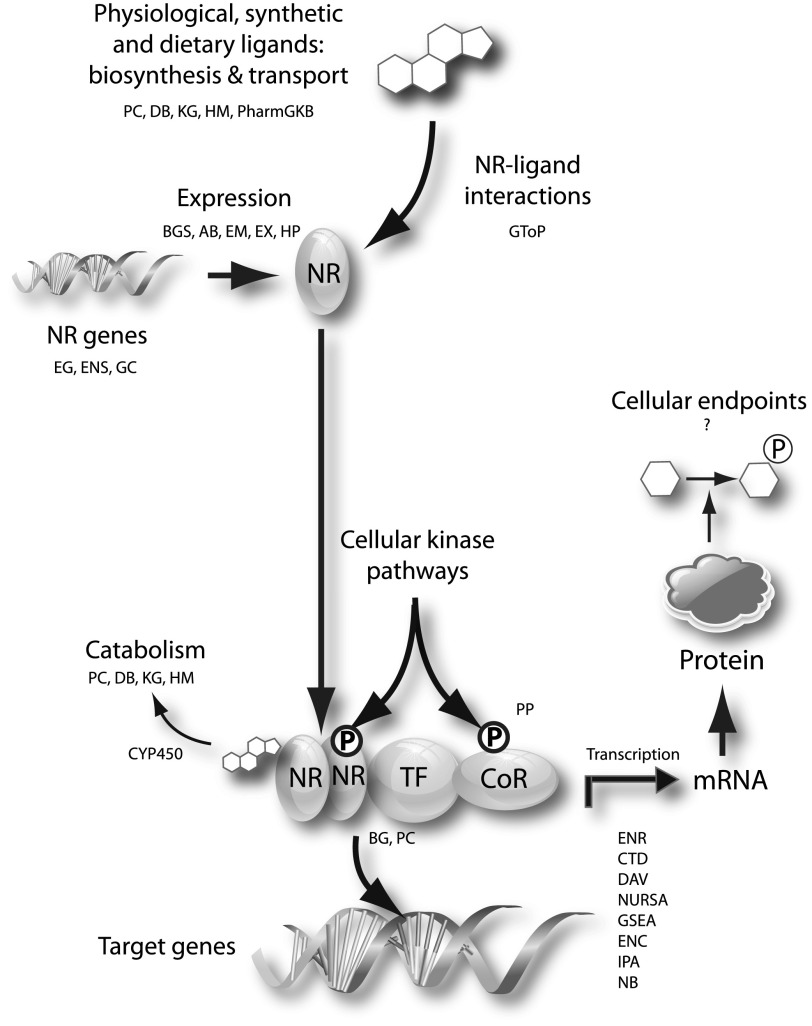
Over the past decade, the field of NR signaling has generated a large volume of global datasets that collectively describe sequences of NR and coregulator genes (genomics): the regulation by NRs and coregulators of gene networks in specific target tissues (transcriptomics); protein-protein interactions and post-translational modifications required for the efficient function of NRs and coregulators (proteomics); specific sites of action of NRs in target gene promoters (cistromics); covalent modification of chromatin (epigenomics); and, more recently, their effects on serum and cellular levels of key metabolites and metabolic intermediates (metabolomics) (Fig. 1). Complementing the efforts of the cell biology community in these areas has been the output of the highly active field of clinical chemistry, which has generated a large number of small molecules to probe the fine details of NR signaling pathway function. A greater appreciation of the tissue-specific pharmacology of NR signaling pathways can be assisted by the availability of web-based tools, free or subscription fee-based, that can be routinely accessed by bench scientists with little or no specialist informatics training. We review below a group of examples of such tools, emphasizing where possible their utility for the pharmacology community. For purposes of comparison, we have defined “signaling pathway” broadly, to encompass the following: metabolism of physiologic and synthetic NR ligands; NR and Cora genes, their expression and their protein products; proteomics, including interactions and post-translational modifications; and selected functional endpoints of NR signaling as described by transcriptomics, genomic DNA-binding analysis, and metabolomics. It is not the intent of this review to critically evaluate each resource or point out shortcomings, but rather to highlight those aspects of each resource that we consider to be most useful to the bench pharmacologist. Table 1 contains URLs and literature references for all of the resources cited in the text. Note that although only one of the resources below—the Nuclear Receptor Signaling Atlas (NURSA)—is a curated NR-centric resource, they all encompass information of relevance to NR pathways.
Fig. 1.
General schematic model of a NR signaling pathway. Abbreviations refer to the web resources listed in Table 1. AB, Allen Brain Atlas; BG, BIOGRID; BGS, BioGPS; CTD, Comparative Toxicogenomics Database; DAV, DAVID; DB, DrugBank; EG, Entrez Gene; EM, Edinburgh Mouse; ENC, ENCODE; ENR, ENRICHR; ENS, Ensembl; EX, Expression Atlas; GC, GeneCards; GSEA, GeneSet Enrichment Analysis; GtoP, International Union of Pharmacology Guide To Pharmacology; HM, Human Metabolite Database; HP, Human Protein Atlas; IPA, Ingenuity; KG, KEGG; NURSA, NURSA Transcriptomine; NB, NextBio; PC, Pathway Commons; PC. PharmGKB, Pharmacogenomics Knowledge Base PubChem; PP, Phosphosite Plus.
TABLE 1.
Web-based resources for exploring NR signaling pathways
| Resource | Name | URL | Reference | ||
|---|---|---|---|---|---|
| Ligands | Physicochemical properties; biosynthesis, transport, and catabolism | PubChem | pubchem.ncbi.nlm.nih.gov | (Kim et al., 2016) | |
| DrugBank | drugbank.ca | (Law et al., 2014) | |||
| KEGG | genome.jp | (Kanehisa et al., 2014) | |||
| HMDB | hmdb.ca | (Wishart et al., 2013) | |||
| Ligand pharmacogenomics | PharmGKB | pharmgkb.org | (Whirl-Carrillo et al., 2012) | ||
| NRs and ligands | Mappings Probes | IUPHAR GtoP | guidetopharmacology.org | (Southan et al., 2016) | |
| NIH Molecular Libraries | http://mli.nih.gov/mli | (Schreiber et al., 2015) | |||
| NRs and CoRs | Genes, transcripts, and proteins | NCBI Entrez Gene | gene.ncbi.nlm.nih.gov | (NCBI Resource Coordinators, 2016) | |
| Ensembl | ensembl.org | (Hubbard et al., 2002) | |||
| UniProt | uniprot.org | (UniProt Consortium, 2014) | |||
| GeneCards | genecards.org | (Safran et al., 2010) | |||
| Expression | BioGPS | biogps.org | (Siragusa et al., 2015) | ||
| Allen Brain Atlas | brain-map.org | (Kuan et al., 2015) | |||
| Edinburgh Mouse Atlas Expression Atlas | emouseatlas.org ebi.ac.uk/gxa | (Richardson et al., 2015; Petryszak et al., 2016) | |||
| Proteomics | Structures Interactions Post-translational modifications | PDB | rcsb.org | (Rose et al., 2015) | |
| BioGRID | biogrid.org | (Chatr-Aryamontri et al., 2015) | |||
| Pathway Commons | pathwaycommons.org | (Cerami et al., 2011) | |||
| Phosphosite Plus | phosphosite.org | (Hornbeck et al., 2015) | |||
| Transcriptomics, cistromics, | CTDBase | ctd.org | (Davis et al., 2015) | ||
| and epigenomics | DAVID | david.ncifcrf.gov | (Sherman et al., 2007) | ||
| NURSA Transcriptomine | nursa.org/transcriptomine | (Becnel et al., 2015) | |||
| GSEA/mSIGDB | broadinstitute.org/gsea/msigdb | (Liberzon et al., 2015) | |||
| ENCODE | encode.gov | (ENCODE Project Consortium, 2004) | |||
| Ingenuity | ingenuity.com | (www.ingenuity.com) | |||
| NextBio | nextbio.com | (www.nextbio.com) | |||
Ligands
General Physicochemical Properties.
A number of excellent general chemical resources document general properties of bioactive small molecules, the most comprehensive being PubChem, ChEBI, and DrugBank. Of particular interest to pharmacology field is the detailed information in these resources on absorption, distribution, and excretion of physiologic NR ligands and the safety and toxicity profiles of synthetic analogs and mimetics. DrugBank contains a particularly comprehensive listing of commercially available forms.
Ligand Biosynthesis, Metabolism, and Pharmacogenomics.
The bioavailability of physiologic and synthetic ER agonists and antagonists is determined in larger part by their cellular and systemic concentrations. Information on the physiologic ligand biosynthetic pathways, encompassing the metabolites, the enzymes, and the genes that encode them, as well as their catabolism, are the manually annotated KEGG, HMDB, and DrugBank resources. A particularly attractive aspect of KEGG is its comprehensive graphical depictions of pathways that highlight the relationships between physiologic NR ligands that go a long way to helping the user understand the key molecular interactions and relationships of these molecules. The ovarian steroidogenesis pathway, for example, which encompasses 17βE2 biosynthesis, is displayed as a visual schematic, with the various biosynthetic intermediates and enzymes represented as nodes that link to contextual information, including information on small-molecule inhibitors of those enzymes. Pharmacogenomic interactions between NR ligands that are approved regulatory drugs and single-nucleotide polymorphisms in human genes encoding their catabolic enzymes are the compass of the Pharmacogenomics KnowledgeBase. Like many of the pre-eminent web-based resources in the field, PharmGKB is based in large part upon manual curation.
NRs and Ligands
The impact of a given small-molecule regulator of NR function on any given NR signaling pathway is defined in part by their affinities for a range of potential NR-binding partners in a tissue. Although a substantial body of literature has been devoted to this discipline, few sites exist to distill these numerous studies into a researcher-accessible form. The most comprehensive public resource, and the only one in existence that actively curates NR-ligand mappings on a consistent basis, is the International Union of Pharmacology’s Guide to Pharmacology. Ligands and receptor mappings, along with essential kinetic information and literature citations, can be found in records in either category of molecule. Mappings of small-molecule NR agonists and antagonists to their cognate receptors can also be found in KEGG and HMDB. Small-molecule perturbants and probes for specific NR signaling pathways, along with information on the assays in which they were screened, are available through the National Institutes of Health’s Molecular Libraries Program.
NRs and Coregulators
Genes, Transcripts, and Proteins.
Another important component of NR signaling pharmacology pathway is the spatiotemporal availability of cognate receptors for small-molecule perturbants. Numerous broad-based gene and protein-centric resources exist that compile, with varying degrees of comprehensive and annotation quality, information on genes, their expression, and the proteins they encode, including National Center for Biotechnology Information Entrez Gene, Ensembl, UniProt, and GeneCards. Of these, GeneCards is, in the author’s opinion, the most comprehensively curated with respect to the various mechanistic, biologic, and clinical aspects of different genes and proteins.
Expression.
A number of resources exist that contain well-curated, systematically collected datasets documenting tissue-specific gene expression patterns of NRs and coregulators (CoRs). Useful expression profiling-based tools for broadly identifying potentially pharmacologically relevant tissues for a given receptor include BioGPS, which takes a specific human, mouse, or rat gene name and returns its relative expression profile across a variety of major tissue types and organs, as well as National Center for Biotechnology Information’s Gene Expression Omnibus and European Bioinformatics Institute’s Expression Atlas. More granular, anatomic resources include Allen Brain Atlas, Human Protein Atlas, and the Edinburgh Mouse Atlas. Allen Brain Atlas in particular represents an impressive undertaking in both the breadth and depth of its coverage and curation, and mapping of its content to gene symbols provides for easier linking with external resources.
Proteomics.
An important aspect of the pharmacology of NR signaling pathways is crosstalk between these pathways and other cell signaling pathways. The molecular events associated with such crosstalk are the purview of proteomics, encompassing protein-protein interactions between NRs, other transcription factors and CoRs, and post-translational modifications of these proteins. Probably the most comprehensive resource in existence for protein interactions is BioGRID, which aggregates information from both high-throughput, discovery-driven datasets as well as low-throughput, hypothesis-driven research articles. A search for estrogen receptor α returned a total of more than 1200 physical interactions extracted from more than 330 publications, categorized according to the original assay method, including coimmunoprecipitation/Western, two-hybrid, and native complex reconstitution. Where available, crystal structures for NR, ligand, and/or CoR interactions are available at the Protein Data Bank, which can be searched either by protein name or by small-molecule perturbant, where one is present in the structure. Protein Data Bank features an attractive user interface and highly detailed curation, and, for many publishers, deposition of crystal coordinates in Protein Data Bank is required as a condition of publication of an article. The pre-eminent resource for post-translational modifications of NRs and CoRs and their coregulators is the manually curated Phosphosite Plus, which documents experimentally demonstrated protein phosphorylation and other post-translational modifications, the conservation of these sites across different species, putative targeting pathways, and links to supporting articles in the literature. Finally, the visually appealing Pathway Commons returns network diagrams indicating relationships between estrogen receptor α and other proteins or genes, curated from the literature.
Transcriptomics, Cistromics, and Epigenomics
The tissue-selective pharmacology of NR signaling pathways is perhaps best understood in terms of the disparity in events downstream of ligand-receptor interactions, principally the following: 1) genomic binding sites of ligand-receptor complexes and 2) regulation of mRNA transcript levels. Transcriptomic analysis of NR signaling pathways involves global-scale relative abundance studies of these events in response to a specific perturbation, such as ligand versus Veh, knockdown, or knockout versus Con or overexpression of a given receptor. The NURSA Transcriptomine database aggregates NR transcriptomic datasets from public archives, supplements their annotation, and organizes them into gene regulation reference libraries for each of the major NR signaling pathways. Access to these libraries is either through individual datasets, linked where possible to their associated primary research articles, or through the Transcriptomine search engine, which allows for customized queries encompassing NR signaling pathway and organ or tissue. Individual data points in a given contrast are linked back to the complete gene list on the dataset page, where related datasets can be discovered, and the dataset can be cited. The ligand/NR/coregulator-gene-tissue/cell line relationships contained in Transcriptomine allow for evidence gathering, hypothesis generation, and model testing by the bench biologist from any background, and assumes no prior familiarity with the field on their part. Examples of datasets associated with articles in Molecular Pharmacology include a study comparing the transcriptomic effects in mouse liver of synthetic agonists of PPAR-α/PPARA (Tijet et al., 2006a,b) and the endocrine-disrupting chemical 2,3,7,8-tetrachlorodibenzodioxin (Kane et al., 2009a,b), an arylhydrocarbon receptor agonist implicated in transcriptional activation of the constitutive androstane receptor (Petrick and Klaassen, 2007).
A similar aggregative approach is adopted by the Comparative Toxicogenomics Database and NextBio. Like NURSA Transcriptomine, Comparative Toxicogenomics Database (CTD) and NextBio assume no prior knowledge of a specific pathway on the part of a user, allowing them to retrieve information concerning the role of NR signaling pathways on regulation of specific gene or group of genes of interest or, conversely, the top regulated genes for a given pathway. Another category of transcriptomic databases, exemplified by Ingenuity Pathway Analysis, ENRICHR, DAVID, and GSEA/MsigDB, is aimed at more sophisticated informatics users. These resources rank the functional similarity of a user-supplied gene list against a set of reference gene lists compiled from public transcriptomic archives to give the user a sense of what signaling pathways are impacted in their perturbations. Broadly speaking, these two categories of tools are complementary, the latter allowing users to gain a broader perspective of the number of pathways impacted in their experimental models, and the former enabling them to drill down on specific genes and pathways in specific tissues. The ENCODE project—the former based upon de novo datasets and the latter on public archives—compiles genome-wide DNA binding and histone modification (chromatin immunoprecipitation) datasets into searchable resources, where DNA-binding transcription factors and source material (cell line or tissue) can be searched and compared with histone modification patterns at specific promoters.
Hypothesis Generation Use Cases
Many data points in discovery-scale or ‘omics datasets are not described in their associated research articles and, as such, are not optimally exposed to search engines such as Google or PubMed or PubMed Central. Accordingly, research resources aggregate and/or annotate these datasets and make them available for data validation and the generation of mechanistic hypotheses in areas of biology that may be new or unfamiliar to their users. In this work, with specific reference to articles in Molecular Pharmacology, we illustrate the use of these resources to create or validate connections between distinct signaling pathways in specific physiologic and pathologic contexts.
Citing decreased expression of the mismatch repair gene MSH2 in oxidatively stressed renal carcinoma cells, Ponnusamy et al. (2016) posited loss of mismatch repair as a potential mechanism for acquired resistance to doxorubicin-induced cytotoxicity in these cells. A search for MSH2 in the Transcriptomine, CTDBase, and NextBio resources found multiple data points documenting its previously uncharacterized repression in liver cells by the xenobiotic phenobarbital (Lambert et al., 2009a,b) and by the GR agonists dexamethasone (Revollo et al., 2013) and methylprednisolone (https://www.nursa.org/nursa/datasets/dataset.jsf?doi=10.1621/SnRGxa6Pww). In contrast, literature searches of SCOPUS, PubMed, and Google failed to identify a relationship between MSH2 and these molecules. These data points support the mechanistic hypothesis that the reported suppression of hepatic apoptosis by phenobarbital (Sanders and Thorgeirsson, 1999) and glucocorticoids (Bailly-Maitre et al., 2001; Oh et al., 2006; Gruver-Yates and Cidlowski, 2013) may be attributable, at least in part, to downregulation of MSH2 expression. Furthermore, activation by dexamethasone of PXR (Pascussi et al., 2000), which in turn is a well-characterized target of phenobarbital (Willson and Kliewer, 2002), suggests positive crosstalk between these pathways in the liver, which is again consistent with their concordant patterns of regulation of MSH2.
A second use case concerns a Molecular Pharmacology article in which Jung et al. postulated that induction of the xenobiotic efflux pump ABCG2 gene by the c-MET/PI3K pathway played a role in the development of chemoresistance in ovarian cancer cells (Jung, 2015 #96). A search for Transcriptomine echoed these findings, showing that ABCG2 was downregulated in gastric cancer cells treated with a c-Met/HGF inhibitor, PHA665-772. In addition to these corroborating data points, Transcriptomine provided evidence postulating previously uncharacterized relationships between ABCG2 and NR signaling pathways. Repression of ABCG2 by the AR/androgen signaling pathway in prostate epithelium LNCaP cells (Hieronymus et al., 2006; Kazmin et al., 2006; Nickols and Dervan, 2007; Nwachukwu et al., 2009; Patki et al., 2013) is consistent with its repression by dihydrotestosterone in breast cancer cells (Chua et al., 2016) and suggests a possible mechanism for the relatively low epithelial expression levels of ABCG2 compared with other prostate cell types (Pascal et al., 2007). Transcriptomine also provided evidence for induction of ABCG2 by 17β-estradiol in osteoblasts (Monroe et al., 2005; Cvoro et al., 2008; Krum et al., 2008; Ball et al., 2009; Paruthiyil et al., 2009). Induction of ABCG2 in bone is consistent with recent reports of the support by 17βE2 of the osteogenic lineage in a variety of stem cell populations (Taskiran and Evren, 2011; Irmak et al., 2014; Li et al., 2014). Moreover, ABCG2 expression was also induced in bone by diarylpropionitrile, which is selective for the ERβ/ESR2, the ER subform that predominates in bone (Paruthiyil et al., 2009). The effects of the ER/estrogen signaling pathway on ABCG2 expression in the bone contrast with its more familiar repression of ABCG2 in mammary gland experimental model systems (Imai et al., 2005) and suggest the testable hypothesis that induction of ABCG2 by the ER/estrogen signaling pathway supports the osteogenic lineage. These examples serve to illustrate the confidence that results when numerous independent datasets cross-validate to postulate a specific gene tissue-signaling pathway relationship.
Concluding Remarks
A broad variety of databases, knowledge bases, and tools exists to support the efforts of bench researchers in the field of NR signaling and its related disciplines. The best of these combine research-focused user interfaces, robust manual curation, and full attribution and acknowledgment of the original studies and their authors. Reviewing them, however, it is difficult to escape the conclusion that they could offer the user so much more if there were better integration between them, such that scientists in one discipline could be readily exposed to information curated from another. This is certainly a complex task, but, given the increasing investment by funding agencies in the infrastructure to support the management of biomedical data, the opportunity is greater now than it has been before. Meaningful integration will require databases to adopt common standards for the exchange of their data, and funding agencies are best placed to ensure that such standards are adopted. These agencies should also ensure that funds are prioritized to support those tools that are both easily locatable by researchers and useful to them. Modern social media seems well placed to allow researchers to provide real-time information on the user base of the various resources—reviews such as this, for example, would be more informative if their authors had access to objective metrics of the impact and utility of these resources in their respective fields. Moreover, improved linkages between journal articles—still the primary means by which researchers consume scientific information—and tools for analyzing the underlying datasets would go a long way toward raising awareness of the number and diversity of resources available to researchers.
As the question mark in Fig. 1 suggests, a notable deficit, for NR and cytoplasmic signaling pathways alike, is the absence of web tools for analyzing their tissue-specific impact on cellular metabolites in normal physiology and metabolic disease. Despite the sharp rise in recent years in signaling metabolomic studies—the number of such studies has increased by more than 1700% over the past decade, compared with an overall growth of signaling articles in PubMed of 75% over the same period—there is currently no freely available resource in which the regulation of cellular metabolism by signal transduction pathways can be compared and contrasted in a user-friendly fashion. Although standards for metabolomics data do exist, albeit in nascent form, deposition of these datasets is yet to be mandated by funding bodies. A brief survey of the recent literature, the details of which are beyond the scope of this review, determined that the deposition of metabolomics datasets in the NR signaling field in public archives is the exception rather than the rule. Given the widely accepted potential of metabolomics to bridge the gap between cell signaling and translational therapeutics (Sreekumar et al., 2009; Hirschey et al., 2010), this seems a missed opportunity for the research community, and there is a collective responsibility on the part of publishers, data repository sponsors, and investigators to redress this situation.
The feudal nature of scientific research and communication—investigators, publishers, funding organizations, citation managers, databases, knowledge bases—complicates attempts to bring order to the often overwhelming number of distinct sources of biomedical data to which the bench researcher is exposed. This complexity has most likely supported the perception held by some that investment in the infrastructure to support data reuse has been misspent and might be better allocated to hypothesis-driven research. In the author’s own experience, however, the deficits in the utility of many tools are attributable largely to the existence in the public domain of many improperly archived and poorly annotated datasets. To address this long-standing issue, funding agencies should consider supporting resources that provide assistance to investigators in the deposition of their datasets in repositories, so as not to burden their research with the relatively mundane, but important, task of depositing the datasets. Equally, community knowledge bases and data repositories should map their curated content to controlled vocabularies and ontologies to support automated and federated distribution, so that their content is visible and leverageable across diverse research communities. Sharing of data holds great promise, but bench scientists will fully embrace data reuse only when those data are freely and easily accessible, comprehensively and accurately annotated, and intuitively presented and integrated with other similar resources. The field of NR signaling looks forward to a new generation of biomedical research resources based on genuine and enduring commitments to these principles.
Abbreviations
- CoR
coregulator
- CTD
C-terminal domain
- ER
estrogen receptor
- NR
nuclear receptor
- NURSA
Nuclear Receptor Signaling Atlas
- PPAR
peroxisome proliferator-activated receptor
Authorship Contributions
Wrote or contributed to the writing of the manuscript: McKenna.
Footnotes
This work was supported by the National Institutes of Health National Institute of Diabetes, Digestive and Kidney Disease and National Institute of Child Health and Development (to the Nuclear Receptor Signaling Atlas) [Grant U24 DK097748].
References
- Bailly-Maitre B, de Sousa G, Boulukos K, Gugenheim J, Rahmani R. (2001) Dexamethasone inhibits spontaneous apoptosis in primary cultures of human and rat hepatocytes via Bcl-2 and Bcl-xL induction. Cell Death Differ 8:279–288. [DOI] [PubMed] [Google Scholar]
- Ball LJ, Levy N, Zhao X, Griffin C, Tagliaferri M, Cohen I, Ricke WA, Speed TP, Firestone GL, and Leitman DC (2009) Analysis of the 17β-estradiol (17βE2)-, raloxifene (Ral)- and Tamoxifen (Tam)-regulated transcriptomes in U2OS cells stably expressing estrogen receptor-α (ERα/ESR1) and ERβ/ESR2. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016.
- Becnel LB, Darlington YF, Ochsner SA, Easton-Marks JR, Watkins CM, McOwiti A, Kankanamge WH, Wise MW, DeHart M, Margolis RN, et al. (2015) Nuclear Receptor Signaling Atlas: opening access to the biology of nuclear receptor signaling pathways. PLoS One 10:e0135615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burris TP, Solt LA, Wang Y, Crumbley C, Banerjee S, Griffett K, Lundasen T, Hughes T, Kojetin DJ. (2013) Nuclear receptors and their selective pharmacologic modulators. Pharmacol Rev 65:710–778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter KD, Korach KS. (2006) Potential biological functions emerging from the different estrogen receptors. Ann N Y Acad Sci 1092:361–373. [DOI] [PubMed] [Google Scholar]
- Cerami EG, Gross BE, Demir E, Rodchenkov I, Babur O, Anwar N, Schultz N, Bader GD, Sander C. (2011) Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res 39:D685–D690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatr-Aryamontri A, Breitkreutz BJ, Oughtred R, Boucher L, Heinicke S, Chen D, Stark C, Breitkreutz A, Kolas N, O’Donnell L, et al. (2015) The BioGRID interaction database: 2015 update. Nucleic Acids Res 43:D470–D478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christakos S, Dhawan P, Verstuyf A, Verlinden L, Carmeliet G. (2016) Vitamin D: metabolism, molecular mechanism of action, and pleiotropic effects. Physiol Rev 96:365–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chua VY, Larma I, Harvey J, Thomas MA, Bentel JM. (2016). Activity of ABCG2 is regulated by its expression and localization in DHT and cyclopamine treated breast cancer cells. J Cell Biochem DOI: 10.1002/jcb.25523 [published ahead of print]. [DOI] [PubMed] [Google Scholar]
- Cvoro A, Tatomer D, Tee MK, Zogovic T, Harris HA, and Leitman DC (2008) Analysis of the 17β-estradiol (17βE2)-regulated transcriptome in the presence of TNFα in U2OS cells stably expressing estrogen receptor-β (ERβ/ESR2). Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/eWxm5Nziy8.
- Davis AP, Grondin CJ, Lennon-Hopkins K, Saraceni-Richards C, Sciaky D, King BL, Wiegers TC, Mattingly CJ. (2015) The Comparative Toxicogenomics Database’s 10th year anniversary: update 2015. Nucleic Acids Res 43:D914–D920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ENCODE Project Consortium (2004) The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306:636–640. [DOI] [PubMed] [Google Scholar]
- Giordano Attianese GM, Desvergne B. (2015) Integrative and systemic approaches for evaluating PPARβ/δ (PPARD) function. Nucl Recept Signal 13:e001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giudici M, Goni S, Fan R, and Treuter E (2016) Nuclear receptor coregulators in metabolism and disease Handb Exp Pharmacol 233:95–135. [DOI] [PubMed] [Google Scholar]
- Gizzo S, Saccardi C, Patrelli TS, Berretta R, Capobianco G, Di Gangi S, Vacilotto A, Bertocco A, Noventa M, Ancona E, et al. (2013) Update on raloxifene: mechanism of action, clinical efficacy, adverse effects, and contraindications. Obstet Gynecol Surv 68:467–481. [DOI] [PubMed] [Google Scholar]
- Glass CK, Ogawa S. (2006) Combinatorial roles of nuclear receptors in inflammation and immunity. Nat Rev Immunol 6:44–55. [DOI] [PubMed] [Google Scholar]
- Glass CK, Rose DW, Rosenfeld MG. (1997) Nuclear receptor coactivators. Curr Opin Cell Biol 9:222–232. [DOI] [PubMed] [Google Scholar]
- Granner DK, Wang JC, Yamamoto KR. (2015) Regulatory actions of glucocorticoid hormones: from organisms to mechanisms. Adv Exp Med Biol 872:3–31. [DOI] [PubMed] [Google Scholar]
- Gruver-Yates AL, Cidlowski JA. (2013) Tissue-specific actions of glucocorticoids on apoptosis: a double-edged sword. Cells 2:202–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hieronymus H, Lamb J, Ross KN, Peng XP, Clement C, Rodina A, Nieto M, Du J, Stegmaier K, Raj SM, et al. (2006) Analysis of the R1881-regulated transcriptome in celastrol-treated LNCaP cells. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/aWNqvAebS2.
- Hirschey MD, Shimazu T, Goetzman E, Jing E, Schwer B, Lombard DB, Grueter CA, Harris C, Biddinger S, Ilkayeva OR, et al. (2010) SIRT3 regulates mitochondrial fatty-acid oxidation by reversible enzyme deacetylation. Nature 464:121–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornbeck PV, Zhang B, Murray B, Kornhauser JM, Latham V, Skrzypek E. (2015) PhosphoSitePlus, 2014: mutations, PTMs and recalibrations. Nucleic Acids Res 43:D512–D520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, Cuff J, Curwen V, Down T, et al. (2002) The Ensembl genome database project. Nucleic Acids Res 30:38–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imai Y, Ishikawa E, Asada S, Sugimoto Y. (2005) Estrogen-mediated post transcriptional down-regulation of breast cancer resistance protein/ABCG2. Cancer Res 65:596–604. [PubMed] [Google Scholar]
- Irmak G, Demirtaş TT, Çetin Altındal D, Çalış M, Gümüşderelioğlu M. (2014) Sustained release of 17β-estradiol stimulates osteogenic differentiation of adipose tissue-derived mesenchymal stem cells on chitosan-hydroxyapatite scaffolds. Cells Tissues Organs 199:37–50. [DOI] [PubMed] [Google Scholar]
- Janani C, Ranjitha Kumari BD. (2015) PPAR gamma gene: a review. Diabetes Metab Syndr 9:46–50. [DOI] [PubMed] [Google Scholar]
- Jung KA, Choi BH, Kwak MK. (2015) The c-MET/PI3K signaling is associated with cancer resistance to doxorubicin and photodynamic therapy by elevating BCRP/ABCG2 expression. Mol Pharmacol 87:465–76. [DOI] [PubMed] [Google Scholar]
- Kane CD, Stevens KA, Fischer JE, Haghpassand M, Royer LJ, Aldinger C, Landschulz KT, Zagouras P, Bagley SW, Hada W, et al. (2009a) Molecular characterization of novel and selective peroxisome proliferator-activated receptor alpha agonists with robust hypolipidemic activity in vivo. Mol Pharmacol 75:296–306. [DOI] [PubMed] [Google Scholar]
- Kane CD, Stevens KA, Fischer JE, Haghpassand M, Royer LJ, Aldinger C, Landschulz KT, Zagouras P, Bagley SW, Hada W, et al. (2009b) Time course analysis of the hypolipidemic peroxisome proliferator activated receptor α (PPARα/Ppara) agonist CP-775146-, CP-868388- or CP-865520-regulated transcriptomes in mouse liver. Nuclear Receptor Signaling Atlas Datasets. Accessed: February 10, 2016. 10.1621/N64DDAoSNK.
- Kanehisa M, Goto S, Sato Y, Kawashima M, Furumichi M, Tanabe M. (2014) Data, information, knowledge and principle: back to metabolism in KEGG. Nucleic Acids Res 42:D199–D205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazmin D, Prytkova T, Cook CE, Wolfinger R, Chu TM, Beratan D, Norris JD, Chang CY, and McDonnell DP (2006) Analysis of the dihydrotestosterone (DHT)- and RTI 6413-018-dependent transcriptomes in LNCaP prostate cancer cells. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/PG6srxDzNt.
- Kim S, Thiessen PA, Bolton EE, Chen J, Fu G, Gindulyte A, Han L, He J, He S, Shoemaker BA, et al. (2016) PubChem Substance and Compound databases. Nucleic Acids Res 44:D1202–D1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krum SA, Miranda-Carboni GA, Lupien M, Eeckhoute J, Carroll JS, and Brown M (2008) Time course transcriptomic analysis of 17β-estradiol (17βE2)-treated U2OS cells stably expressing estrogen receptor-α (ERα/ESR1). Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/3RnoklZ5K1.
- Kuan L, Li Y, Lau C, Feng D, Bernard A, Sunkin SM, Zeng H, Dang C, Hawrylycz M, Ng L. (2015) Neuroinformatics of the Allen Mouse Brain Connectivity Atlas. Methods 73:4–17. [DOI] [PubMed] [Google Scholar]
- Lambert CB, Spire C, Renaud MP, Claude N, and Guillouzo A (2009a) Analysis of the phenobarbital (Phenb)-regulated transcriptome in HepaRG cells. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/RdJpfZvFp6.
- Lambert CB, Spire C, Renaud MP, Claude N, Guillouzo A. (2009b) Reproducible chemical-induced changes in gene expression profiles in human hepatoma HepaRG cells under various experimental conditions. Toxicol In Vitro 23:466–475. [DOI] [PubMed] [Google Scholar]
- Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al. (2014) DrugBank 4.0: shedding new light on drug metabolism. Nucleic Acids Res 42:D1091–D1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Yan M, Wang Z, Zheng Y, Li J, Ma S, Liu G, Yu J. (2014) 17beta-Estradiol promotes the odonto/osteogenic differentiation of stem cells from apical papilla via mitogen-activated protein kinase pathway. Stem Cell Res Ther 5:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. (2015) The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1:417–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. (1995) The nuclear receptor superfamily: the second decade. Cell 83:835–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mark M, Ghyselinck NB, Chambon P. (2009) Function of retinoic acid receptors during embryonic development. Nucl Recept Signal 7:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, Lanz RB, O’Malley BW. (1999) Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev 20:321–344. [DOI] [PubMed] [Google Scholar]
- Monroe DG, Secreto FJ, Subramaniam M, Getz BJ, Khosla S, and Spelsberg TC (2005) Comparative analysis of estrogen receptor-α (ERα/ESR1) or ERβ/ESR2-dependent transcriptomes in 17β-estradiol (17βE2)- or 4HT-treated U2OS cells. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/4ZXZjH7niR.
- Mustonen MV, Pyrhönen S, Kellokumpu-Lehtinen PL. (2014) Toremifene in the treatment of breast cancer. World J Clin Oncol 5:393–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NCBI Resource Coordinators (2016) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 44:D7–D19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickols NG and Dervan PB (2007) Analysis of the androgen response element-independent and AR-independent transcriptomes in dihydrotestosterone (DHT)-treated LNCaP cells. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/Y2NYDQA5ZI.
- Nwachukwu JC, Mita P, Ruoff R, Ha S, Wang Q, Huang SJ, Taneja SS, Brown M, Gerald WL, Garabedian MJ, and Logan SK (2009) Analysis of the UXT and R1881 dependent transcriptomes in LNCaP prostate cancer cells. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/gZmIbonYZ2.
- Oh HY, Namkoong S, Lee SJ, Por E, Kim CK, Billiar TR, Han JA, Ha KS, Chung HT, Kwon YG, et al. (2006) Dexamethasone protects primary cultured hepatocytes from death receptor-mediated apoptosis by upregulation of cFLIP. Cell Death Differ 13:512–523. [DOI] [PubMed] [Google Scholar]
- Paruthiyil S, Cvoro A, Zhao X, Wu Z, Sui Y, Staub RE, Baggett S, Herber CB, Griffin C, Tagliaferri M, et al. (2009) Analysis of the 17β-estradiol (17βE2)- and diarylpropionitrile (DPN)-regulated transcriptomes in U2OS cells stably expressing estrogen receptor-α (ERα/ESR1)- and ERβ/ESR2. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/rGRCXFd8iw.
- Pascal LE, Oudes AJ, Petersen TW, Goo YA, Walashek LS, True LD, Liu AY. (2007) Molecular and cellular characterization of ABCG2 in the prostate. BMC Urol 7:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascussi JM, Drocourt L, Fabre JM, Maurel P, Vilarem MJ. (2000) Dexamethasone induces pregnane X receptor and retinoid X receptor-alpha expression in human hepatocytes: synergistic increase of CYP3A4 induction by pregnane X receptor activators. Mol Pharmacol 58:361–372. [DOI] [PubMed] [Google Scholar]
- Patki M, Chari V, Sivakumaran S, Gonit M, Trumbly R, and Ratnam M (2013) Analysis of the ELK and R1881-dependent transcriptomes in LNCaP prostate cancer cells. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/xZngfuhLwA.
- Petrick JS, Klaassen CD. (2007) Importance of hepatic induction of constitutive androstane receptor and other transcription factors that regulate xenobiotic metabolism and transport. Drug Metab Dispos 35:1806–1815. [DOI] [PubMed] [Google Scholar]
- Petryszak R, Keays M, Tang YA, Fonseca NA, Barrera E, Burdett T, Füllgrabe A, Fuentes AM, Jupp S, Koskinen S, et al. (2016) Expression Atlas update: an integrated database of gene and protein expression in humans, animals and plants. Nucleic Acids Res 44:D746–D752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinkerton JV, Stanczyk FZ. (2014) Clinical effects of selective estrogen receptor modulators on vulvar and vaginal atrophy. Menopause 21:309–319. [DOI] [PubMed] [Google Scholar]
- Ponnusamy L, Mahalingaiah PK, Singh KP. (2016) Chronic oxidative stress increases resistance to doxorubicin-induced cytotoxicity in renal carcinoma cells potentially through epigenetic mechanism. Mol Pharmacol 89:27–41. [DOI] [PubMed] [Google Scholar]
- Pyper SR, Viswakarma N, Yu S, Reddy JK. (2010) PPARalpha: energy combustion, hypolipidemia, inflammation and cancer. Nucl Recept Signal 8:e002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Revollo JR, Oakley RH, Lu NZ, Kadmiel M, Gandhavadi M, and Cidlowski JA (2013) Analysis of the dexamethasone (Dex)-regulated, hairy and enhancer of split-1 (Hes1)-dependent murine hepatic transcriptome. Nuclear Receptor Signaling Atlas Datasets. Accessed: April 7, 2016. 10.1621/mbbL1izhup.
- Richardson L, Graham L, Moss J, Burton N, Roochun Y, Armit C, Baldock RA. (2015) Developing the eHistology Atlas. Database (Oxford) DOI:10.1093/database/bav105. [DOI] [PMC free article] [PubMed]
- Rose PW, Prlić A, Bi C, Bluhm WF, Christie CH, Dutta S, Green RK, Goodsell DS, Westbrook JD, Woo J, et al. (2015) The RCSB Protein Data Bank: views of structural biology for basic and applied research and education. Nucleic Acids Res 43:D345–D356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubel CA, Jeong JW, Tsai SY, Lydon JP, Demayo FJ. (2010) Epithelial-stromal interaction and progesterone receptors in the mouse uterus. Semin Reprod Med 28:27–35. [DOI] [PubMed] [Google Scholar]
- Safe S, Jin UH, Hedrick E, Reeder A, Lee SO. (2014) Minireview: role of orphan nuclear receptors in cancer and potential as drug targets. Mol Endocrinol 28:157–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al. (2010) GeneCards Version 3: the human gene integrator. Database (Oxford) 2010:baq020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders S, Thorgeirsson SS. (1999) Phenobarbital promotes liver growth in c-myc/TGF-alpha transgenic mice by inducing hypertrophy and inhibiting apoptosis. Carcinogenesis 20:41–49. [DOI] [PubMed] [Google Scholar]
- Schreiber SL, Kotz JD, Li M, Aubé J, Austin CP, Reed JC, Rosen H, White EL, Sklar LA, Lindsley CW, et al. NIH Molecular Libraries Project Team (2015) Advancing biological understanding and therapeutics discovery with small-molecule probes. Cell 161:1252–1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulman IG. (2010) Nuclear receptors as drug targets for metabolic disease. Adv Drug Deliv Rev 62:1307–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman BT, Huang W, Tan Q, Guo Y, Bour S, Liu D, Stephens R, Baseler MW, Lane HC, Lempicki RA. (2007) DAVID Knowledgebase: a gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinformatics 8:426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siragusa L, Cross S, Baroni M, Goracci L, Cruciani G. (2015) BioGPS: navigating biological space to predict polypharmacology, off-targeting, and selectivity. Proteins 83:517–532. [DOI] [PubMed] [Google Scholar]
- Soccio RE, Chen ER, Lazar MA. (2014) Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab 20:573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southan C, Sharman JL, Benson HE, Faccenda E, Pawson AJ, Alexander SP, Buneman OP, Davenport AP, McGrath JC, Peters JA, et al. NC-IUPHAR (2016) The IUPHAR/BPS Guide to PHARMACOLOGY in 2016: towards curated quantitative interactions between 1300 protein targets and 6000 ligands. Nucleic Acids Res 44:D1054–D1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreekumar A, Poisson LM, Rajendiran TM, Khan AP, Cao Q, Yu J, Laxman B, Mehra R, Lonigro RJ, Li Y, et al. (2009) Metabolomic profiles delineate potential role for sarcosine in prostate cancer progression. Nature 457:910–914. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- Taskiran D, Evren V. (2011) Stimulatory effect of 17β-estradiol on osteogenic differentiation potential of rat adipose tissue-derived stem cells. Gen Physiol Biophys 30:167–174. [DOI] [PubMed] [Google Scholar]
- Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, and Pohjanvirta R (2006a) Analysis of the arylhydrocarbon receptor (Ahr)-regulated hepatic transcriptome in 2,3,7,8-tetrachlorodibenzodioxin (TCDD)-treated mice. Nuclear Receptor Signaling Atlas Datasets. Accessed: February 10, 2016. 10.1621/P9XIQWjG4I.
- Tijet N, Boutros PC, Moffat ID, Okey AB, Tuomisto J, Pohjanvirta R. (2006b) Aryl hydrocarbon receptor regulates distinct dioxin-dependent and dioxin-independent gene batteries. Mol Pharmacol 69:140–153. [DOI] [PubMed] [Google Scholar]
- UniProt Consortium (2014) Activities at the Universal Protein Resource (UniProt). Nucleic Acids Res 42:D191–D198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whirl-Carrillo M, McDonagh EM, Hebert JM, Gong L, Sangkuhl K, Thorn CF, Altman RB, Klein TE. (2012) Pharmacogenomics knowledge for personalized medicine. Clin Pharmacol Ther 92:414–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willson TM, Kliewer SA. (2002) PXR, CAR and drug metabolism. Nat Rev Drug Discov 1:259–266. [DOI] [PubMed] [Google Scholar]
- Wishart DS, Jewison T, Guo AC, Wilson M, Knox C, Liu Y, Djoumbou Y, Mandal R, Aziat F, Dong E, et al. (2013) HMDB 3.0: The Human Metabolome Database in 2013. Nucleic Acids Res 41:D801–D807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C, Dahlman-Wright K, Gustafsson JA. (2008) Estrogen receptor beta: an overview and update. Nucl Recept Signal 6:e003. [DOI] [PMC free article] [PubMed] [Google Scholar]