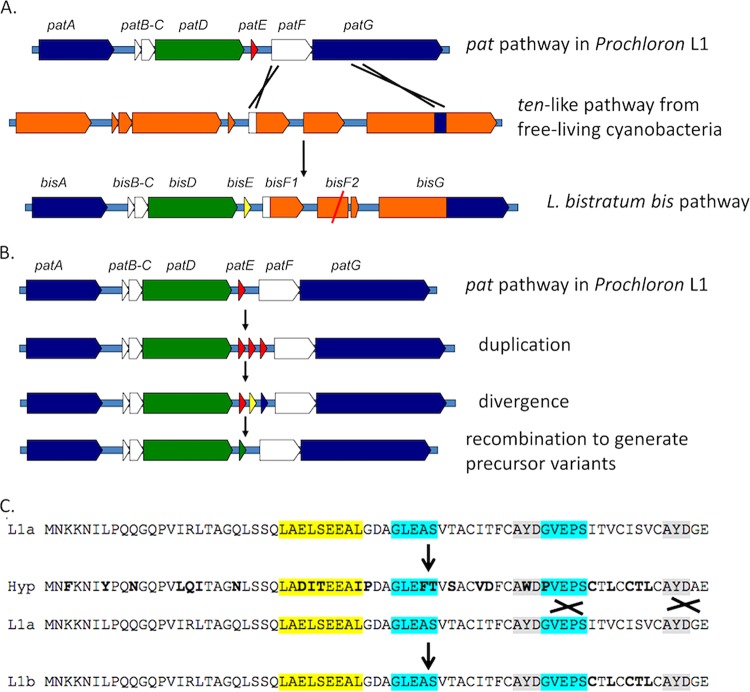
FIG 7.
Evolutionary model of cyanobactin biosynthesis. Because cyanobactin enzymes exhibit DG properties of broad-substrate tolerance, genetic swapping (likely recombination) leads to generation of new variants. Our model is as follows. (A) Capture of new functional enzymes. The pat pathway captured a ten-like cluster, possibly from free-living cyanobacteria, leading to the ability to produce hexapeptides (only heptapeptides and octapeptides are known from pat-like clusters). Subsequently, a second copy of the F gene was degraded into a pseudogene via point mutation. In panels A and B, colors are meant to make it easier to see the origin of genes but have no functional meaning. (B) Evolution of new precursor peptides. Precursor peptides likely evolve based upon duplication, divergence, and recombination. One way that this could work is shown here, with the duplication of precursors within a pathway based upon what is known from other cyanobactin pathways (18). This enables ready generation of sequence variants. (C) Example of how duplicated sequences might enable generation of precise variants for DG metabolism. Protein sequence is shown in place of gene sequence for ease of comprehension. Sequences L1a and L1b are real sequences from L. patella L1, while Hyp is a hypothetical, diverged intermediate. Important residues for recognition sequences I, II, and III are shown in yellow, blue, and gray, respectively.