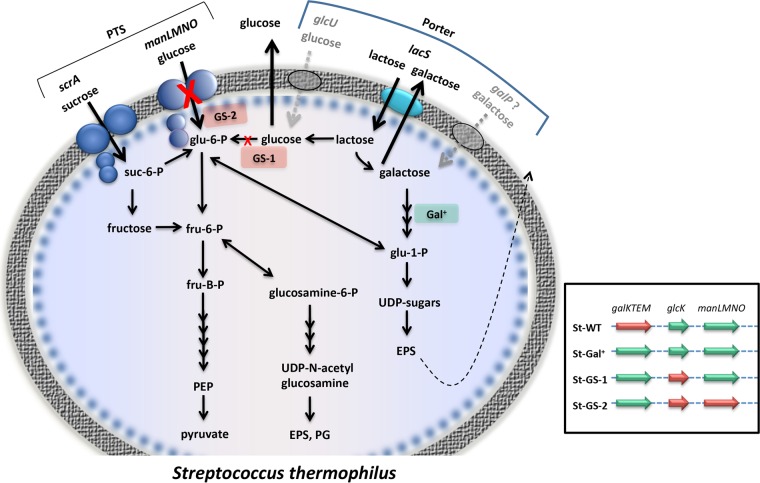
FIG 3.
Sugar metabolism in S. thermophilus. Black arrows, active enzyme reactions; dotted gray arrows, putative enzyme reactions; red Xs, enzyme reactions inactivated by mutations; dotted black arrow, externalization of exopolysaccharide. The indicated genes are as follows: scrA, sucrose PTS component II; manLMNO, glucose/mannose PTS IIABCD; lacS, lactose permease; glcU, glucose permease; and galP, putative galactose permease. PTS, phosphotransferase system; EPS, exopolysaccharide; PG, peptidoglycan; PEP, phosphoenolpyruvate; glu-1-P, glucose-1-phosphate; glu-6-P, glucose-6-phosphate; fru-6-P, fructose-6-phosphate; fru-B-P, fructose-1,6-biphosphate; suc-6-P, sucrose-6-phosphate. (Inset) Green arrows, active genes; red arrows, inactive genes. Genes are as follows: galKTEM, galactose operon; glcK, glucokinase; manLMNO, and glucose/mannose PTS IIABCD.