ABSTRACT
Investigating the physiology of cyanobacteria cultured under a diel light regime is relevant for a better understanding of the resulting growth characteristics and for specific biotechnological applications that are foreseen for these photosynthetic organisms. Here, we present the results of a multiomics study of the model cyanobacterium Synechocystis sp. strain PCC 6803, cultured in a lab-scale photobioreactor in physiological conditions relevant for large-scale culturing. The culture was sparged with N2 and CO2, leading to an anoxic environment during the dark period. Growth followed the availability of light. Metabolite analysis performed with 1H nuclear magnetic resonance analysis showed that amino acids involved in nitrogen and sulfur assimilation showed elevated levels in the light. Most protein levels, analyzed through mass spectrometry, remained rather stable. However, several high-light-response proteins and stress-response proteins showed distinct changes at the onset of the light period. Microarray-based transcript analysis found common patterns of ∼56% of the transcriptome following the diel regime. These oscillating transcripts could be grouped coarsely into genes that were upregulated and downregulated in the dark period. The accumulated glycogen was degraded in the anaerobic environment in the dark. A small part was degraded gradually, reflecting basic maintenance requirements of the cells in darkness. Surprisingly, the largest part was degraded rapidly in a short time span at the end of the dark period. This degradation could allow rapid formation of metabolic intermediates at the end of the dark period, preparing the cells for the resumption of growth at the start of the light period.
IMPORTANCE Industrial-scale biotechnological applications are anticipated for cyanobacteria. We simulated large-scale high-cell-density culturing of Synechocystis sp. PCC 6803 under a diel light regime in a lab-scale photobioreactor. In BG-11 medium, Synechocystis grew only in the light. Metabolite analysis grouped the collected samples according to the light and dark conditions. Proteome analysis suggested that the majority of enzyme-activity regulation was not hierarchical but rather occurred through enzyme activity regulation. An abrupt light-on condition induced high-light-stress proteins. Transcript analysis showed distinct patterns for the light and dark periods. Glycogen gradually accumulated in the light and was rapidly consumed in the last quarter of the dark period. This suggests that the circadian clock primed the cellular machinery for immediate resumption of growth in the light.
INTRODUCTION
Understanding cyanobacterial physiology in a diel environment is of interest to understand circadian regulation in general and for the utilization of these organisms in biotechnological applications. Our exploration of the effect of a diel light cycle on a cyanobacterial culture started with the wish to investigate the response of the metabolic network of the cells to the imposed repetitively fluctuating environment, considering future biotechnological applications. The attractiveness of employing a photosynthetic organism in industrial biotechnology is that it can harness the energy from the sun, even though this energy is available only in a dilute and intermittent fashion. Utilizing cyanobacteria for large-scale product formation will benefit from a high cell density, provided the trade-off of self-shading stays within reasonable bounds. Industrial-scale cultures of cyanobacteria growing in large-scale closed photobioreactors are envisioned to be fed by CO2-rich exhaust gases from combustion engines and/or anaerobic digesters. Such culturing conditions may result in a microaerobic or an anoxic environment when photosynthesis stops in darkness.
Most biotechnological applications of cyanobacteria are based on the use of synthetic biology to reroute intermediary metabolism, such that heterologous production pathways allow formation of a preferred product (1). Intermediary metabolism forms a highly interconnected network, initiated by nutrient influx and governed by multiple layers of regulation. Whether engineered metabolic routes result in high rates of product formation depends on the metabolic flux capacities, the proper regulatory signals, and the availability of the intracellular substrate(s). Hence, the productivity of cyanobacteria is highly dependent on the changes of their metabolite levels and/or regulation of metabolism during the daily cycle (2–5). Rhythms of the metabolism are affected by the light-dark transitions and also by the circadian clock, both with distinct effects (6). Disentangling those influences will benefit the optimization of cyanobacterial production systems. Cyanobacteria, such as Synechocystis sp. strain PCC 6803 (here, Synechocystis) are oxygenic photoautotrophic microorganisms that can be modified to produce a wide range of products (7–9). Accordingly, CO2 can be invested, via the cyanobacterial metabolism, as the carbon substrate for sustainable production of biofuels, bulk chemicals, etc., thus generating CO2-neutral energy carriers and commodity chemicals (10). Consequently, there is an interest in the large-scale growth of these organisms, in the compounds contained in their biomass, and in compounds excreted into the growth medium (11). So far, excreted products, synthesized via heterologous pathways, typically are precursors to bioplastics (e.g., butane-diol and lactic acid) (12, 13), biofuels (e.g., ethanol and butanol) (14), and secondary metabolites and flavor compounds, such as vitamins and terpenes (15, 16).
In this study, wild-type Synechocystis cells were grown in continuous culture in a turbidostat-controlled lab-scale photobioreactor, sparged with a mixture of N2 and CO2 (17). The culture was subjected to a diel rhythm of 12 h of light and 12 h of darkness (LD), which resulted in rapid oxygen depletion during the dark period. These are conditions that mimic the conditions of mass culturing in a large-scale closed photobioreactor, equipped with a degassing system and sparged with off-gases originating from the combustion of fossil fuel (18) or an anaerobic digester (19). To achieve high cell densities, high light intensities are needed which, depending on geographical location, may regularly exceed the photosynthetic capacity of cells (especially at midday in full sunlight). Such a changing environment, specifically with respect to the primary source of energy (light) and the lack of oxygen in the dark period, may have a strong influence on the metabolic state and metabolic efficiency of cyanobacteria; the organism, for instance, will have to react to the anoxic environment by fermentation to cover the energy requirement for cellular maintenance (3, 20).
To understand these physiological responses in more detail, we set out to investigate the metabolome, proteome, and transcriptome of such cells from samples taken at time points close to the changes of light availability to study the immediate response and the subsequent adaptation to the diel regime. The most striking observations that were made will be discussed in detail.
MATERIALS AND METHODS
Strain and preculture conditions.
A preculture of the glucose-tolerant wild-type strain of Synechocystis sp. PCC 6803 (strain obtained from D. Bhaya, Stanford) was incubated in BG-11 medium (Sigma, St. Louis, MO, USA) and grown at 30°C in a shaking incubator at 120 rpm (Innova 43; New Brunswick Scientific, Enfield, CT, USA). Cultures were illuminated with constant moderate light provided by 15 W of cool fluorescent white lights (F15T8-PL/AQ; General Electric, CT, USA). The resulting light intensity was 30 μmol photons/m2/s measured with a LI-250 light meter (Li-Cor, Lincoln, NE, USA). Prior to the experiments in the photobioreactor (see below), a 100-ml preculture was cultivated in BG-11 medium in a 300-ml Erlenmeyer flask. To ensure that an axenic culture was used, an aliquot of the culture was spread on BG-11 plates and on LB plates (solidified with 1.5%, wt/vol, agar). Plates were incubated at 30°C and screened for 3 consecutive days for potential contaminants. BG-11 plates were supplemented with 10 mM TES buffer at pH 8 {N-[Tris(hydroxymethyl)methyl]-2-aminoethanesulfonic acid adjusted to the relevant pH with KOH}, 5 mM glucose, and 0.3% sodium thiosulfate.
Photobioreactor.
The Synechocystis preculture (see above) was used to seed two FMT-150 photobioreactors (PBRs) (Photon System Instruments, Brno, Czech Republic) with 100 ml of culture each. This study makes use of the ∼1-liter vessel model FMT-150 that is temperature controlled and illuminated from one side by blue- and red-light emitting diodes (LEDs) (17). Growth was monitored by the integrated densitometer as the optical density measured at 735 nm (OD735). The culture medium was BG-11 (Sigma). Continuous gas mixing was provided by the gas mixing system GMS150 (Photon System Instruments, Brno, Czech Republic) set to 0.5% CO2/99.5% N2, coupled to mass flow controllers (Smart Mass Flow Model 5850S; Brooks Instruments, PA, USA) to supply 0.5 liters/min to each PBR. The CO2 influx provided an excess of (inorganic) carbon and clamped the pH in the preferred range of 7.5 to 8.0. The temperature was kept at 30°C ± 1°C. The lid of the PBRs accommodated a pH electrode, a Clark-type concentration of dissolved oxygen (dO2) electrode, and a potentiometric concentration of dissolved carbon dioxide (dCO2) electrode (Mettler-Toledo). The light regime applied by the LED board provided 12 h of illumination and 12 h of darkness in square-wave cycles (LD). The culture was kept at an OD735 of 0.8 ± 0.01, as measured by the integrated densitometer at 735 nm and regulated by the turbidostat control. This value corresponded to ∼2.57 ± 0.05 determined at OD730 in a benchtop spectrophotometer with 1-cm path length (Lightwave II; Biochrom, Cambridge, United Kingdom), and this was kept constant over multiple consecutive days of the LD regime (see Fig. S1 to S4 in the supplemental material). The pump coupled to the turbidostat control attached to the medium reservoir was set to 5.6 ml/min, resulting in a saw-tooth pattern in the OD reading from the repetitive influx of medium when the upper bound of the threshold was reached until the medium pump was stopped again at the lower bound. The slope between the pump intervals was used to determine the growth rate. The two vessels were illuminated with LED arrays consisting of blue (445 nm, 18-nm full width at half maximum [FWHM]; and 458 nm, 16-nm FWHM, respectively) and red (for both PBRs: 636 nm, 20-nm FWHM) LEDs (17). The LED boards provided 500 μmol photons/m2/s of each wavelength to the culture vessels. Less than 5% of the light input was measured at the opposite side of each vessel, probably implying a light-limited growth regime for the whole population (21). After an initial period of continuous illumination, to reach the target cell density rapidly (i.e., in a couple of days), the LD regime was applied over a period of 1 week until a quasi-steady state was reached. Quasi-steady state was judged from the optical density measurements and visual inspection of the recurring patterns of the OD735 and the OD680, the ratio of which was used as a measure for chlorophyll content (see Fig. S2 in the supplemental material). Next, over a period of 10 consecutive days, samples were taken for metabolite analysis. Thereafter, within 3 consecutive 24-h periods, samples for protein analysis, glycogen determination, and dry cell weight (gram dry weight [gDW]) determinations were taken (for sampling time points, see Fig. 1; see also Fig. S1 in the supplemental material). The repetitive signal-output patterns of the two PBR systems (OD735, OD680, dO2, dCO2, pH, and temperature) were essentially identical for the whole duration of the experiment (see Fig. S2 to S4). The dO2 and the OD680 signal showed slightly decreasing amplitude during the initial 1-week period that was used to establish a quasi-steady state. This signal stabilized, however, before the sampling took place (see Fig. S2A and S3). For growth rate determination, a best fit based on an exponential equation was derived from the OD735 signal, between the preset thresholds for the turbidostat control, which controlled the supply of fresh medium (see Fig. S2). For the identification of the first actual growth phase after the shift to the light period, the lower threshold effectively indicated the end of the lag phase. It should be noted that the rapid drop of the OD735 signal at the onset of the dark period was an artifact of the optical density measurement caused by the temperature change of the equipment due to the switching off of the light (data not shown). Likewise, the rapid increase of the OD735 signal at the onset of illumination was caused by the reverse effect.
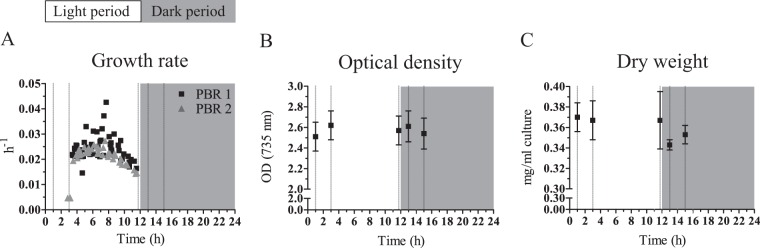
FIG 1.
Growth characteristics of a turbidostat-controlled culture treated with a repetitive diel light regime of 12 h of light and 12 h of dark. Dashed vertical lines represent the time points of sampling for dry weight, metabolite, protein, and transcript analysis. (A) Growth rates, calculated by linear regression of log-transformed OD735, change during the growth phases in the light. Data from multiple days (1 week) for both PBRs are shown. Depending on the starting OD at the beginning of each light period, this resulted in 5 to 9 consecutive pumping/nonpumping intervals. The respective x axis value was chosen from the end time point of the time window of the nonpumping interval. (B) Average optical density showing a stable signal at the dedicated sampling time points. Values are the average and standard deviation derived from the biological replicates (n = 2). (C) The cell dry weight shows a rather stable value. Values are the average and standard deviation derived from the biological replicates (n = 2) and two technical replicates (n = 2). Note the split y axes for B and C.
For the metabolite analysis, a 50-ml cell suspension was harvested for each nuclear magnetic resonance (NMR) sample (∼18 mg dry weight). Sampling 5 times per 24-h period resulted in the removal of 250 ml of culture (from a total of 1 liter), which still allowed a stable growth regime over multiple subsequent days (see Fig. S2 to S4 in the supplemental material). About 270 to 300 ml of new medium was pumped in during each 24-h period, which, under this chosen regime, compensated for the removal of culture and allowed for the continuous culturing. We selected 9 (PBR1) and 10 (PBR2) consecutive days for NMR sampling to have enough replicates for the statistical analysis.
Glycogen analysis.
Rapid sampling from the PBRs for glycogen determination was performed as follows: about 5 ml of cell suspension from the vessel was harvested through a sampling port with a syringe into a 15-ml tube. For samples from the dark period, the tubes were covered with aluminum foil. Aliquots of a 2-ml cell suspension (technical duplicates for each PBR) were pelleted by centrifugation at 14,000 rpm and 4°C for 5 min (in a prechilled centrifuge). The supernatant was removed carefully, and the remaining wet pellet was stored at −20°C for batch processing at a later time point. Glycogen was determined essentially as described before (22), employing the d-fructose/d-glucose assay kit (Megazyme) adapted for use in a 96-well plate reader. Cell pellets were resuspended in 200 μl of KOH (5.35 M) and hydrolyzed for 90 min at 95°C in a thermomixer at 500 rpm. For glycogen precipitation, 600 μl of cold ethanol (absolute, Scharlau) was added to previously cooled samples and placed on ice for 2 h. The insoluble glycogen was pelleted by centrifugation at 14,000 rpm at 4°C for 5 min (in a prechilled centrifuge) and washed twice with cold ethanol. The remaining pellet was supplemented with 300 μl of acetate buffer (200 mM, pH 5.2) and 50 μl of amyloglucosidase (Roche) dissolved in acetate buffer and incubated overnight at 55°C under constant agitation. Samples were centrifuged at 5,000 rpm at room temperature for 1 min, and the glucose concentration was determined with the d-fructose/d-glucose assay kit (Megazyme) according to the manufacturer instructions. The amount of glycogen was normalized to the gDW.
Dry cell weight measurements.
For the determination of the dry cell weight for each PBR, two subsequent aliquots of 25 ml of the culture were harvested into the same 30-ml preweighted glass tubes. The cell suspensions were pelleted by centrifugation at 10,000 rpm at room temperature for 10 min. The supernatant was removed carefully without disturbing the pellets, after which the cell pellets were dried overnight in a stove at 110°C. The tubes were subsequently weighted.
High-performance liquid chromatography analysis of supernatant samples.
We analyzed the external medium of the culture for potentially excreted fermentation products. Given a total volume of 1 liter for the culture in the PBRs, which had 0.36 ± 0.01 gDW (Fig. 1C), 5.1% of this (the maximal difference in glycogen content of the cells) (see Fig. 4B) was 18 mg. Hence, 18 mg of a C6 compound (such as glucose, the building block of the polymer glycogen) could have been mobilized from the internal glycogen storage. If invested into a C6 excretion product (for example, glucose), we could expect a concentration of 0.1 mM in the extracellular medium. However, in supernatant samples from the 5 time points, we did not detect any of the common fermentation products, such as lactic acid, acetic acid, formic acid, etc.; all of these products have a detection limit of 50 μM on our high-performance liquid chromatography system (data not shown). This was tested as follows: 1 ml of supernatant samples was treated with 100 μl of 35% perchloric acid (Merck), incubated on ice for 10 min, and neutralized with 50 μl of 7M KOH (Merck). After thorough mixing, the precipitate was removed by centrifugation for 2 min at 12,000 rpm and filtered (minisart SRP4, 0.45 μm; Sartorius Stedin Biotech). Separation of organic acids and alcohols was achieved with a Rezex ROA-organic acid H+ (8%) column (Phenomenex) at 45°C using a flow of 0.5 ml/min. A refractive index detector (RI-1530; Jasco) and AZUR 4.5 Software (Datalys) were used for detection.
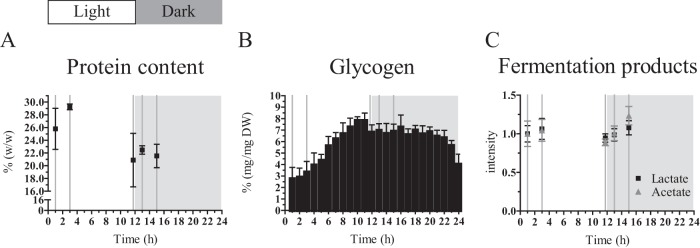
FIG 4.
Cellular composition of a turbidostat-controlled culture treated with a diel light regime of 12 h of light and 12 h of dark. Dashed vertical lines represent the time points of sampling for dry weight, metabolite, protein, and transcript analysis. (A) The protein content of the cells is higher at early time points of the light period compared to the later time point and compared to the samples from the dark period. Values are expressed in percentage of dry weight. Values are the average and standard deviation derived from the biological replicates (n = 2) and two technical replicates (n = 2). Note the split y axis. (B) The glycogen content shows a distinct accumulation and degradation pattern. Values are expressed in percentage of dry weight. Values are the average and standard deviation derived from the biological replicates (n = 2) and two technical replicates (n = 2). (C) Metabolite analysis reveals a constant concentration of intracellular lactate and a slight increase for intracellular acetate in the early dark period. Intensity values are normalized to the value of the first sampling time point in the light period. Mean and standard error of the mean are shown.
Accession number(s).
Proteomic data were deposited in the PRIDE database under accession number PXD001619.
RESULTS AND DISCUSSION
A multiomics approach was used to characterize the cellular physiology of Synechocystis over a diel light cycle in a quasi-steady state. Such a quasi-steady state, which refers to no change from cycle to cycle, was obtained by growing Synechocystis in a photobioreactor (PBR) with turbidostat control with 12 h of illumination and 12 h of darkness. The PBR enabled automatic acquisition of growth data through the integrated densitometer measuring optical density (OD) at 680 and 735 nm, as well as frequent sampling for glycogen content. For metabolite, transcriptome, and proteome analysis (see Materials and Methods in the supplemental material) as well as sampling for dry cell weight and protein content, we chose to focus sampling around the time point of the shift from the light period to the dark period. This was motivated by the findings of Kucho et al. (23), which indicated significant changes in gene expression (thus indicating hierarchical regulation) at the transition from subjective day to subjective night under continuous-light conditions in an entrained culture.
Growth is dynamic and exclusive to the light period.
The slopes in OD735, arising from growth between the pump intervals (see Fig. S1 in the supplemental material), allow determination of the growth rate in the respective time windows (Fig. 1A). With the exception of some outliers, the growth rate varied between 0.015 and 0.030 h−1 (which corresponded to doubling times of ∼40 and 20 h, respectively), following a distinct pattern over the light period. After the dark period and a lag phase without growth (see Fig. S1), the first time window that allowed a growth rate determination showed a growth rate of ∼0.020 h−1. The growth rate then increased over the course of the light period, peaked early in the second half of the light period (∼8 h after light onset), and decreased toward its end, at which point the growth rate had decreased to 0.015 h−1. Thus, the growth rate showed an arc-like shape as a function of time in the 12-h light period, even though the light intensity and the OD735 (and dry weight) were constant during this time (Fig. 1B and C). This dynamic growth rate was certainly expected for an experimental setup, as presented for Synechocystis in, e.g., Labiosa et al. (24), where a dimming schedule to simulate an actual diel cycle (achieved by sinusoidal illumination) was used. However, a similar pattern for the growth rate arose here, despite our use of square-wave light-dark cycles. Therefore, although we did not simulate light availability and intensity according to natural diel settings, we observed the same growth behavior, suggesting that corresponding physiological parameters could be comparable to past studies. Similarly, though phase advanced with respect to the growth rate, the OD680-to-OD735 ratio, which is indicative of chlorophyll content, increased and subsequently decreased over the course of the day (see Fig. S2B). This oscillation can be explained by the circadian clock, which Synechocystis is equipped with (25, 26). We have recently shown that this pattern of growth rate and chlorophyll content also occurs in an entrained culture in continuous-light conditions (27).
Metabolite, proteome, and transcript data place nitrogen and sulfur assimilation into the light period and indicate hierarchical regulation.
To see whether the observed pattern of growth rates was reflected in the acquired omics data, we used statistical analysis to evaluate the different omics profiles. Multivariate data analysis employing a supervised method, partial least square discriminant analysis of the metabolite signals from the 5 time points collected (in Fig. 1, the dashed vertical lines represent the time points of sampling), showed significant changes of the metabolite profile over the 24-h period (see Fig. S5 in the supplemental material). Next, we aimed at finding proteins that changed in time and shared a biological function, annotated in the cyanobacterial sequence information repository Cyanobase (28). By performing analysis of variance (ANOVA) on the set of all quantified proteins (see Table S1 and proteomic data deposited in PRIDE under accession number PXD001619), we identified 45 proteins that showed a significant change in at least one of the five consecutive time points (P < 0.05) (see Table S2). We refered to those 45 proteins as the changing proteins. The results from the enrichment analysis for a biological function of this set can be found in Table S3. The ANOVA-based transcript analysis of the microarray data revealed that approximately 56% of the genes probed showed a time-dependent change (P < 0.05) (see Table S4).
If both a metabolite and the corresponding enzyme changed in parallel with its gene transcript, hierarchical control could be assumed, whereas, if a metabolite changed without a change in the corresponding protein level, metabolic control could be assumed for the altered flux through the pathway under consideration. Based on the variable importance in the projection (VIP) scores (see Fig. S6 and Table S5 in the supplemental material), the metabolites that showed the largest changes were compared to the proteins involved in their respective metabolism, though the overlap between the metabolites identified (see Table S5) and the proteins quantified (see Table S1) was small. This comparison showed that, from glutamate and glutamine metabolism, which are among the most dynamic metabolites in this study, two enzymes have been identified in the proteome analysis of this study: the glutamine synthases GlnA (Slr1756) and GlnN (Slr0288). GlnA is also present in the list of 45 changing proteins (see Table S2) (P = 0.020), and its transcript is in the list of significantly changing genes (see Table S4) (P = 0.007), showing strong downregulation in the dark (see entry slr1756 in Table S4). Inspection of the GlnA protein levels at the sampling time points showed that it decreased by ∼20% during the course of the light period and remained at the lower level in the dark period, whereas GlnN protein levels showed no significant change (see entry Slr0288 in Table S1). GlnN is mainly induced under nitrogen limiting conditions, explaining the low, stable levels observed (29). GlnA catalyzes the conversion reaction from glutamate to glutamine, thereby branching off intermediates from the tricarboxylic acid cycle. The downregulation of both the GlnA protein levels and the transcript levels correlated with the trend of decreasing glutamine levels in the dark period, and this enzyme might thus be regulated at least partially in a hierarchical fashion. However, glutamate showed reduced levels as well (Fig. 2), suggesting a generally lower influx into the glutamate and glutamine metabolism, implying that nitrogen assimilation took place predominantly in the light.
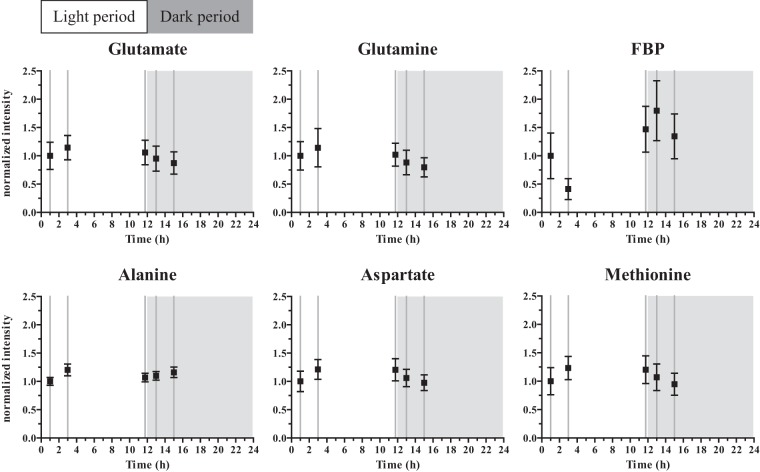
FIG 2.
Dynamically changing metabolites over the light-dark cycle. Selected identified metabolites from the NMR analysis. Intensity values are normalized to the value of the first sampling time point in the light period. Mean and standard error of the mean are shown.
In addition to glutamate and glutamine, methionine showed a similar pattern, with higher levels in the light period (Fig. 2). This pattern was also reflected in aspartate, a precursor to methionine and part of cyanophycin, a known nitrogen storage compound. Methionine is one of the key amino acids through which sulfur is assimilated. When considering the machinery for sulfate reduction, it became apparent that this pathway was significantly downregulated in the night (slr1165, slr0676, slr1791) (P < 0.05) or shows no significant change (slr0963) (P > 0.05) (see Table S4 in the supplemental material). The only protein of that particular pathway that was detected in our study, Slr0676, showed no significant change (P > 0.05). Rather than concluding that this pattern is a general trend of synthesis of all amino acids, it should be noted that some other amino acids that were detected, e.g., glycine and threonine, did not show significant change over time and were among the lower-scoring metabolites (VIP score, <1) (see Table S5 in the supplemental material). One metabolite that did change significantly over time, but did not mimic the trend displayed by the amino acids involved in nitrogen and sulfur assimilation, was alanine. This deviating pattern may have stemmed from multiple pathways in which alanine is involved, such as transamination and, indirectly, glycolysis (30).
Integration of metabolite and proteome data suggests metabolic control for the majority of the Calvin-Benson-Bassham cycle proteins.
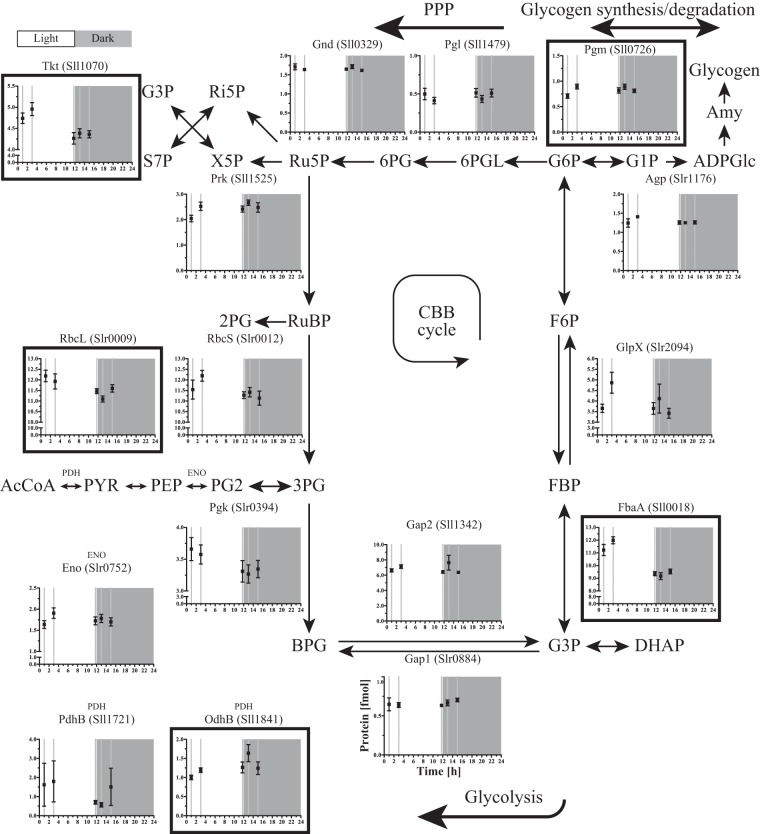
The list of most changing metabolites also contains fructose-1,6-bisphosphate (FBP) (see Table S5 in the supplemental material). FBP showed lower levels at the early time points of the light period, followed by higher levels at later time points, even before the light period ended (Fig. 2) and coinciding with reaching the maximal glycogen content (Fig. 1). FbaA, the FBP aldolase, participating in the Calvin-Benson-Bassham (CBB) cycle, was high at time point 1 and time point 2 and decreased to a lower level again in the 3 remaining sampling time points (Fig. 3), thus showing pattern opposite that of FBP (Fig. 2). FbaA also showed a significant time-dependent change determined by ANOVA (P = 2.0 × 10−8), grouping it with the 45 changing proteins (see Table S2). FBP is a part of both the CBB cycle and the glycolytic pathway (Fig. 3). The amount of Gap2, the prime glyceraldehyde-3-phosphate dehydrogenase in the fixation-direction of the CBB cycle, was about 10-fold higher than the amount of Gap1, which, in turn, has been shown to be mainly active in glycolysis (31). Both proteins seemed to be kept at rather stable levels during the light-dark cycle that was imposed on the cells in this study. The enzyme catalyzing the preceding carbon-incorporating step in the CBB cycle, ribulose-1,5-bisphosphate carboxylase/oxygenase (RuBisCO, composed of the RuBisCO large subunit [RbcL] and the RuBisCO small subunit [RbcS]), showed slightly larger amounts during the early light period and a lower (∼10%) amount in the 3 remaining sampling time points, close to and in the dark period. Of the CBB cycle (Fig. 3), next to FbaA, only RbcL also appeared in the list of changing proteins (P = 0.024). The enzymes of the CBB cycle that are shared with the pentose phosphate pathway (PPP) did not show a significantly altered pattern (Fig. 3). Nevertheless, the fluctuating FBP levels in concordance with the glycogen accumulation/degradation levels (see below) corroborated that the direction of the carbon flow through the regeneration phase of the CBB cycle, which overlaps with glycolysis, inverted between light and dark conditions (32). Here, we showed that this inversion of the direction of the flux only marginally made use of hierarchical regulation of the participating enzymes (Fig. 3). In a recent study of Osanai et al., the transition from aerobic light to anaerobic dark conditions was investigated, with a focus on metabolites (33). Of all metabolites investigated, exactly one, FBP, overlapped with those that we identified here. Interestingly, and despite differences in experimental design, Osanai et al. also found relatively low FBP levels in the light and high levels under anaerobic dark conditions.
FIG 3.
Protein profiles of the CBB and central carbon metabolism. Those proteins that changed significantly over time are marked with a black frame and are referred to as those that are subjected to hierarchical regulation. Mean and standard error of the mean are shown.
Glycogen gradually accumulated in the light and was degraded in two distinct phases in the dark period.
Over the course of a diel cycle, the biomass composition of the cells was not as constant as the OD730 and dry weight (compare Fig. 1B and C). Protein content (the soluble fraction) (see Materials and Methods) was higher in the early than in the late light period and in the (early) dark period (Fig. 4A). An almost opposite pattern could be observed for the glycogen content of the cells (Fig. 4B). Glycogen is the main carbon storage compound of cyanobacteria (34, 35). Here, the glycogen content of the cells changed between 2.7% ± 1.0% and 7.8% ± 0.4% of the dry weight. The lowest levels of glycogen were present at the onset of the light period, but renewed accumulation started immediately after the transition. In the simulated afternoon, the glycogen level reached a maximum and thereafter declined slightly during the rest of the remaining light period, as well as during the majority of the dark period. Remarkably, ∼2 h before the onset of the next light period, rapid glycogen degradation took place, suggesting the investment of carbon into intracellular building blocks for the machinery supporting growth, to prime the cell for the upcoming light period. Based on flux balance analysis of a genome-wide model of the Synechocystis metabolism, Knoop et al. (36) calculated a linear degradation rate for glycogen over the majority of the dark period. Interestingly, that model further predicted that the glycogen content also decreased slightly faster during the last hours of the night. The calculated flux through the participating enzymes in that flux balance analysis showed a higher flux through phosphoglucomutase exactly in the late dark period. Significantly, we did not detect any major fermentation product(s) in the extracellular medium (see Materials and Methods). Nevertheless, we do suggest that Synechocystis fermented in the dark and anoxic period, which led to intracellular acetate and lactate formation. Interestingly, only acetate levels were increased 1 and 3 h after the shift to the dark, whereas lactate levels remained constant over the five sampling points (Fig. 4C). This minimal fermentation activity was apparently not enough to support growth during the dark anoxic phase, when respiration is not possible. We attributed the mild but gradual decrease in glycogen content during the majority of the dark period (before the rapid degradation set in; see above) to its conversion into acetate. The energy yield of this conversion presumably covered the general costs of cellular maintenance during the dark period. It should be noted that this dark, anaerobic phase did not represent a major loss of carbon. The carbon freed from glycogen during the slow degradation phase (i.e., 1.5% [wt/wt] glycogen, between the maximum in the subjective afternoon and the time point just before the onset of the rapid degradation phase) corresponded to 0.0076 mmol carbon/gDW/h, taking glycogen as a C6 compound. This equals 0.1 nmol carbon/1 mg cell protein/min, which was lower than the lowest reported rate of glycogen consumption in cyanobacteria by Stal and Moezelaar (20). This then also set an upper limit to the maintenance requirements of the organism under anaerobic conditions in the dark of <0.038 mmol ATP/gDW/h, assuming complete fermentation to acetate (20). Because of the acetate formation observed, it is likely that reduction of H+ or nitrate/nitrite was used to maintain redox balance. This is consistent with the strong upregulation (top 2% to 12% of most-changing transcripts observed) in the dark of the hox genes (hoxE, hoxF, hoxU, hoxY, and hoxH, which are sll1220, sll1221, sll1223, sll1224, and sll1226) that encode the bidirectional NiFe-hydrogenase, as well as hydrogenase maturation gene hypA1 (slr1675, top 1% of changing transcripts) (see also Table S4 in the supplemental material). Interestingly, the largest overall fold change was observed for sll0741, encoding the pyruvate flavodoxin/ferredoxin oxidoreductase, which was recently shown to be the electron source of the hydrogenase (37). However, protein levels of ferredoxin-nitrite reductase remained stable, whereas ferredoxin-nitrate reductase was not detected (see Table S1). Furthermore, the transcript abundance indicated that nitrate/nitrite transport (sll1450, sll1451, sll1452, and sll1453) and reduction (ferredoxin-nitrite reductase slr0898 and ferredoxin-nitrate reductase sll1454) were downregulated in the dark (see Table S4), which is consistent with our tentative conclusion that hydrogenase was the main redox valve in the dark (see above).
Consequences of square-wave diel cycles.
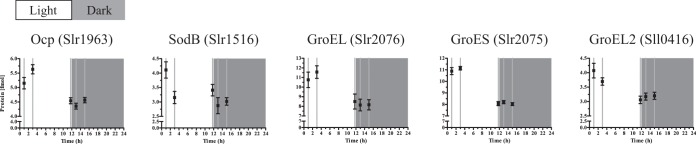
The abrupt transition from dark to high light may cause high-light stress, as suggested by the lag phase in growth of ∼2 h. Proteome analysis revealed that significantly changing proteins are strongly enriched in the class of the high-light and stress-response proteins and chaperones (P < 0.005) (see Table S3 in the supplemental material). Other indicators of stress are expression patterns of the orange carotenoid protein (OCP), the superoxide dismutase (SodB), and the group I chaperonins (GroEL, GroES, and GroEL2) (Fig. 5), which all showed the highest levels at the beginning of the light period, coinciding with the lag phase before growth resumed (see Fig. S1). Significantly, this high-light response was not evident in the gene-expression study of Synechocystis by Labiosa et al. (24), in which light intensity was gradually increased and decreased in time. In addition to the notion of possible initial high-light stress, the cells appeared to experience constant high light during the light period. This followed from the detection of the high-light variant of the D1 protein, PsbA2, at all time points (see entries for Slr1311 in Table S1). Furthermore, the psbA2 (slr1311) and psbA3 (sll1867) transcripts were both very abundant and were downregulated in the dark period, whereas the psbA1 (slr1181) transcript was weakly expressed and was upregulated in the dark period (see Table S4). Accordingly, the significantly changing proteins were enriched in proteins of the category photosystem II (see Table S3). Additionally, the high-light transcript of the D2 gene, psbD2 (slr0927 in Table S4), accumulated to levels similar to those of psbD1 (sll0849). However, chlorophyll content, represented by the OD680-to-OD735 ratio, actually increased in the first 3 h of the light period (see Fig. S2B), and further increasing light intensity did not cause the typical photoinhibition phenotype of gradually reducing PSII activity, which would have led to decreasing oxygen evolution, indicating that the chosen conditions did not exceed the intrinsic photorepair capacity (see Fig. S7). It is important to consider that the used light intensity did not surpass maximum light intensity at noon at moderate latitude and that cell density was high, which led to a steep gradient of light intensity within the culture and an actually fairly low light intensity per cell on average. This is emphasized by the low light transmittance of the culture (∼5% of incident light intensity) and the lower oxygen evolution at the target density than at a lower density.
FIG 5.
High-light-response proteins and stress-response proteins show distinct changes at the onset of the light period. Mean and standard error of the mean are shown. Note the split y axis.
Although abrupt light-dark transitions do not reflect natural progression of light intensity over the course of a day, they facilitate discrimination of a direct light response from circadian regulation. We performed k-means clustering on the ANOVA-filtered set of genes, with k = 10 to detect significantly differing time-dependent change (see Fig. S8 in the supplemental material). The k-means clustering on the set of the changing genes resulted in roughly two types with distinct patterns: the clusters 1, 5, 6, and 8 showed upregulation upon the transition to the dark period, whereas the clusters 2, 3, 4, 7, 9, and 10 showed downregulation. However, cluster 7 was already downregulated before the transition, whereas cluster 8 showed the opposite. In similar fashion, clusters 1 and 4 showed a delay in upregulation and downregulation, respectively. Together, these four clusters represented 443 genes, or ∼13% of the total number of genes probed. Enrichment analysis on the 10 clusters was done to identify significantly enriched functional categories for Synechocystis, based on Cyanobase (see Tables S8 and S9). It is interesting to note that cluster 8 was enriched in genes involved in the PPP, despite clear downregulation of genes involved in RNA and DNA synthesis, which were enriched in cluster 9.
For the set of changing proteins, k-means clustering resulted in four clusters with distinct patterns (see Fig. S9 in the supplemental material). Two clusters, 2 and 4, showed high levels in the early time points, followed by a decrease during the light period, and were both enriched in chaperones (see Table S10 and S11). Cluster 3, on the other hand, showed its lowest levels around the end of the lag phase and was strongly enriched in proteins involved in photosynthesis and respiration. Cluster 1, despite appearing to correlate with light availability, was not significantly enriched in any biological category.
As mentioned above, most of the proteins detected showed little or no changes over the five different time points of the light-dark regime. The proteins with very high P values (above 0.8) from the ANOVA constituted the set of proteins which failed to show any evidence of time-dependent change at all. This prompted the identification of 19 proteins which we thus considered stable over time during the diel regime (see Table S6 in the supplemental material). An enrichment analysis based on the categories of the biological functions showed that phycobilisome proteins (P = 0.006) were among these (see Table S7). The phycobilisome proteins are the most abundant proteins found in the cells (38), which is corroborated by our analysis, and represent a major investment of resources for the cell, explaining their stable levels. It is interesting to note that the phycobilisome proteins showed no change, whereas the apparent chlorophyll content changed over the course of the light period in a fashion similar to content in a culture exposed to a much lower light intensity (see Fig. S2B) (27). We note that the mass spectrometry analysis and subsequent evaluation of the patterns of change were performed after normalization to the total soluble protein content.
Conclusion.
Here, we showed that Synechocystis, in a diel light regime with anaerobic conditions during the dark period, can grow only in the light and maintains a constant dry weight throughout the day. During the light period, glycogen accumulates as protein content decreases, and this reverses at the end of the dark period, restarting the daily cycle. Glycogen, assimilated during the day to a maximum of ∼7.5% dry weight, is gradually fermented for maintenance requirements in the first 10 h of the dark period, followed by rapid degradation in the last 2 h. Nitrogen and sulfur assimilation takes place predominantly in the light, and hydrogen evolution is preferred over nitrate reduction during fermentation in the night. We conclude that maintenance costs are extremely low and require only a small fraction of the glycogen. The later rapid degradation of glycogen prepares the cells for growth in the upcoming light period, though a short lag phase is observed despite this priming of metabolism. Mass culturing under the conditions we have used in this study presumably will thus not be hampered by a huge loss of carbon and energy in the dark period. The use of square-wave diel cycles allows for discriminating between circadian regulation and a direct light response. Our quantified proteome data set and the normalized transcriptome data set facilitate further investigation of potential proteins/genes of interest. The determination of their correlations will benefit the optimization of cyanobacterial production systems.
Supplementary Material
ACKNOWLEDGMENTS
We thank Naira Quintana (presently at Rousselot, Belgium) for the initiative at the 10th Cyanobacterial Molecular Biology Workshop (CMBW), June 2010, Lake Arrowhead, Los Angeles, CA, USA, to start the collaborative endeavor reported here. We thank Timo Maarleveld from CWI/VU (Amsterdam) for a custom-made Python script handling the output from the NMR analysis and for evaluating and visualizing the separate metabolites for their evaluation. We thank Rob Verpoorte from Leiden University (metabolome analysis) and Hans Aerts from the AMC (proteome analysis) for lab space and equipment. We thank Robert Lehmann (Humboldt University Berlin) and Ilka Axmann (University of Düsseldorf) for sharing the R-code for the LOS transformation of the transcript data. We thank Hans C. P. Matthijs from IBED for inspiring dialogues and insightful thoughts on continuous culturing of cyanobacteria. We thank Sandra Waaijenborg for performing the transcript normalization and Johan Westerhuis from BDA, Jeroen van der Steen and Filipe Branco dos Santos from MMP, and Lucas Stal from IBED/NIOZ for helpful discussions. We thank Milou Schuurmans from MMP for help with sampling and glycogen determination. We thank the members of the RNA Biology & Applied Bioinformatics group at SILS, in particular Selina van Leeuwen, Elisa Hoekstra, and Martijs Jonker, for the microarray analysis. We thank the reviewers of this work for their insightful comments which improved the quality of the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00256-16.
REFERENCES
- 1.Quintana N, Van der Kooy F, Van de Rhee MD, Voshol GP, Verpoorte R. 2011. Renewable energy from cyanobacteria: energy production optimization by metabolic pathway engineering. Appl Microbiol Biotechnol 91:471–490. doi: 10.1007/s00253-011-3394-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Niederholtmeyer H, Wolfstädter BT, Savage DF, Silver PA, Way JC. 2010. Engineering cyanobacteria to synthesize and export hydrophilic products. Appl Environ Microbiol 76:3462–3466. doi: 10.1128/AEM.00202-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McNeely K, Xu Y, Ananyev G, Bennette N, Bryant DA, Dismukes GC. 2011. Synechococcus sp. strain PCC 7002 nifJ mutant lacking pyruvate:ferredoxin oxidoreductase. Appl Environ Microbiol 77:2435–2444. doi: 10.1128/AEM.02792-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lan EI, Liao JC. 2011. Metabolic engineering of cyanobacteria for 1-butanol production from carbon dioxide. Metab Eng 13:353–363. doi: 10.1016/j.ymben.2011.04.004. [DOI] [PubMed] [Google Scholar]
- 5.Xu Y, Weyman PD, Umetani M, Xiong J, Qin X, Xu Q, Iwasaki H, Johnson CH. 2013. Circadian yin-yang regulation and its manipulation to globally reprogram gene expression. Curr Biol CB 23:2365–2374. doi: 10.1016/j.cub.2013.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Diamond S, Jun D, Rubin BE, Golden SS. 2015. The circadian oscillator in Synechococcus elongatus controls metabolite partitioning during diurnal growth. Proc Natl Acad Sci U S A 112:E1916–E1925. doi: 10.1073/pnas.1504576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gudmundsson S, Nogales J. 2015. Cyanobacteria as photosynthetic biocatalysts: a systems biology perspective. Mol Biosyst 11:60–70. doi: 10.1039/C4MB00335G. [DOI] [PubMed] [Google Scholar]
- 8.Oliver JWK, Atsumi S. 2014. Metabolic design for cyanobacterial chemical synthesis. Photosynth Res 120:249–261. doi: 10.1007/s11120-014-9997-4. [DOI] [PubMed] [Google Scholar]
- 9.Angermayr SA, Gorchs Rovira A, Hellingwerf KJ. 2015. Metabolic engineering of cyanobacteria for the synthesis of commodity products. Trends Biotechnol 33:352–361. doi: 10.1016/j.tibtech.2015.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Savakis P, Hellingwerf KJ. 2015. Engineering cyanobacteria for direct biofuel production from CO2. Curr Opin Biotechnol 33:8–14. doi: 10.1016/j.copbio.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 11.Wijffels RH, Kruse O, Hellingwerf KJ. 2013. Potential of industrial biotechnology with cyanobacteria and eukaryotic microalgae. Curr Opin Biotechnol 24:405–413. doi: 10.1016/j.copbio.2013.04.004. [DOI] [PubMed] [Google Scholar]
- 12.Savakis PE, Angermayr SA, Hellingwerf KJ. 2013. Synthesis of 2,3-butanediol by Synechocystis sp. PCC6803 via heterologous expression of a catabolic pathway from lactic acid and enterobacteria. Metab Eng 20:121–130. doi: 10.1016/j.ymben.2013.09.008. [DOI] [PubMed] [Google Scholar]
- 13.Angermayr SA, Paszota M, Hellingwerf KJ. 2012. Engineering a cyanobacterial cell factory for production of lactic acid. Appl Environ Microbiol 78:7098–7106. doi: 10.1128/AEM.01587-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ducat DC, Way JC, Silver PA. 2011. Engineering cyanobacteria to generate high-value products. Trends Biotechnol 29:95–103. doi: 10.1016/j.tibtech.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Abed RMM, Dobretsov S, Sudesh K. 2009. Applications of cyanobacteria in biotechnology. J Appl Microbiol 106:1–12. doi: 10.1111/j.1365-2672.2008.03918.x. [DOI] [PubMed] [Google Scholar]
- 16.Pattanaik B, Lindberg P. 2015. Terpenoids and their biosynthesis in cyanobacteria. Life (Basel) 5:269–293. doi: 10.3390/life5010269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nedbal L, Trtílek M, Cervený J, Komárek O, Pakrasi HB. 2008. A photobioreactor system for precision cultivation of photoautotrophic microorganisms and for high-content analysis of suspension dynamics. Biotechnol Bioeng 100:902–910. doi: 10.1002/bit.21833. [DOI] [PubMed] [Google Scholar]
- 18.Janssen M, Tramper J, Mur LR, Wijffels RH. 2003. Enclosed outdoor photobioreactors: light regime, photosynthetic efficiency, scale-up, and future prospects. Biotechnol Bioeng 81:193–210. doi: 10.1002/bit.10468. [DOI] [PubMed] [Google Scholar]
- 19.Converti A, Oliveira RPS, Torres BR, Lodi A, Zilli M. 2009. Biogas production and valorization by means of a two-step biological process. Bioresour Technol 100:5771–5776. doi: 10.1016/j.biortech.2009.05.072. [DOI] [PubMed] [Google Scholar]
- 20.Stal LJ, Moezelaar R. 1997. Fermentation in cyanobacteria. FEMS Microbiol Rev 21:179–211. doi: 10.1016/S0168-6445(97)00056-9. [DOI] [Google Scholar]
- 21.Huisman J, Matthijs HCP, Visser PM, Balke H, Sigon CAM, Passarge J, Weissing FJ, Mur LR. 2002. Principles of the light-limited chemostat: theory and ecological applications. Antonie Van Leeuwenhoek 81:117–133. doi: 10.1023/A:1020537928216. [DOI] [PubMed] [Google Scholar]
- 22.Parrou JL, François J. 1997. A simplified procedure for a rapid and reliable assay of both glycogen and trehalose in whole yeast cells. Anal Biochem 248:186–188. doi: 10.1006/abio.1997.2138. [DOI] [PubMed] [Google Scholar]
- 23.Kucho K, Okamoto K, Tsuchiya Y, Nomura S, Nango M, Kanehisa M, Ishiura M. 2005. Global analysis of circadian expression in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 187:2190–2199. doi: 10.1128/JB.187.6.2190-2199.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Labiosa RG, Arrigo KR, Tu CJ, Bhaya D, Bay S, Grossman AR, Shrager J. 2006. Examination of diel changes in global transcript accumulation in Synechocystis (cyanobacteria). J Phycol 42:622–636. doi: 10.1111/j.1529-8817.2006.00217.x. [DOI] [Google Scholar]
- 25.Aoki S, Kondo T, Ishiura M. 1995. Circadian expression of the dnaK gene in the cyanobacterium Synechocystis sp. strain PCC 6803. J Bacteriol 177:5606–5611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aoki S, Kondo T, Wada H, Ishiura M. 1997. Circadian rhythm of the cyanobacterium Synechocystis sp. strain PCC 6803 in the dark. J Bacteriol 179:5751–5755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Alphen P, Hellingwerf KJ. 2015. Sustained circadian rhythms in continuous light in Synechocystis sp. PCC6803 growing in a well-controlled photobioreactor. PLoS One 10:e0127715. doi: 10.1371/journal.pone.0127715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nakao M, Okamoto S, Kohara M, Fujishiro T, Fujisawa T, Sato S, Tabata S, Kaneko T, Nakamura Y. 2010. CyanoBase: the cyanobacteria genome database update 2010. Nucleic Acids Res 38:D379-381. doi: 10.1093/nar/gkp915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reyes JC, Muro-Pastor MI, Florencio FJ. 1997. Transcription of glutamine synthetase genes (glnA and glnN) from the cyanobacterium Synechocystis sp. strain PCC 6803 is differently regulated in response to nitrogen availability. J Bacteriol 179:2678–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steinhauser D, Fernie AR, Araújo WL. 2012. Unusual cyanobacterial TCA cycles: not broken, just different. Trends Plant Sci 17:503–509. doi: 10.1016/j.tplants.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 31.Koksharova O, Schubert M, Shestakov S, Cerff R. 1998. Genetic and biochemical evidence for distinct key functions of two highly divergent GAPDH genes in catabolic and anabolic carbon flow of the cyanobacterium Synechocystis sp. PCC 6803. Plant Mol Biol 36:183–194. [DOI] [PubMed] [Google Scholar]
- 32.Pelroy RA, Bassham JA. 1972. Photosynthetic and dark carbon metabolism in unicellular blue-green algae. Arch Mikrobiol 86:25–38. doi: 10.1007/BF00412397. [DOI] [PubMed] [Google Scholar]
- 33.Osanai T, Shirai T, Iijima H, Nakaya Y, Okamoto M, Kondo A, Hirai MY. 2015. Genetic manipulation of a metabolic enzyme and a transcriptional regulator increasing succinate excretion from unicellular cyanobacterium. Front Microbiol 6:1064. doi: 10.3389/fmicb.2015.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gründel M, Scheunemann R, Lockau W, Zilliges Y. 2012. Impaired glycogen synthesis causes metabolic overflow reactions and affects stress responses in the cyanobacterium Synechocystis sp. PCC 6803. Microbiology 158:3032–3043. doi: 10.1099/mic.0.062950-0. [DOI] [PubMed] [Google Scholar]
- 35.Zilliges Y. 2014. Glycogen, a dynamic cellular sink and reservoir for carbon, p 189–210. In Flores E, Herrero A (ed), The cell biology of cyanobacteria. Caister Academic Press, Norfolk, United Kingdom. [Google Scholar]
- 36.Knoop H, Gründel M, Zilliges Y, Lehmann R, Hoffmann S, Lockau W, Steuer R. 2013. Flux balance analysis of cyanobacterial metabolism: the metabolic network of Synechocystis sp. PCC 6803. PLoS Comput Biol 9:e1003081. doi: 10.1371/journal.pcbi.1003081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutekunst K, Chen X, Schreiber K, Kaspar U, Makam S, Appel J. 2014. The bidirectional NiFe-hydrogenase in Synechocystis sp. PCC 6803 is reduced by flavodoxin and ferredoxin and is essential under mixotrophic, nitrate-limiting conditions. J Biol Chem 289:1930–1937. doi: 10.1074/jbc.M113.526376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kudoh K, Kawano Y, Hotta S, Sekine M, Watanabe T, Ihara M. 2014. Prerequisite for highly efficient isoprenoid production by cyanobacteria discovered through the overexpression of 1-deoxy-d-xylulose-5-phosphate synthase and carbon allocation analysis. J Biosci Bioeng 118:20–28. doi: 10.1016/j.jbiosc.2013.12.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.