ABSTRACT
Livestock-associated methicillin-resistant Staphylococcus aureus (LA-MRSA) was detected in 2003 and rapidly became the predominant MRSA clade in the Netherlands. Studies have shown that transmissions are difficult to identify, since this MRSA variant represents a genetically homogenous clade when current typing techniques are used. Here, next-generation sequencing was performed on 206 LA-MRSA isolates to assess the capability of LA-MRSA to be transmitted between humans. The usefulness of single nucleotide variants (SNVs), the composition of the SCCmec region, and the presence of plasmids to identify transmission of LA-MRSA were assessed. In total, 30 presumed putative nosocomial transmission events and 2 LA-MRSA outbreaks were studied; in most cases, SNV analysis revealed that the isolates of the index patient and the contact(s) clustered closely together. In three presumed events, the isolates did not cluster together, indicating that transmission was unlikely. The composition of the SCCmec region corroborated these findings. However, plasmid identification did not support our SNV analysis, since different plasmids were present in several cases where SNV and SCCmec analysis suggested that transmission was likely. Next-generation sequencing shows that transmission of LA-MRSA does occur in Dutch health care settings. Transmission was identified based on SNV analysis combined with epidemiological data and in the context of epidemiologically related and unrelated isolates. Analysis of the SCCmec region provided limited, albeit useful, information to corroborate conclusions on transmissions, but plasmid identification did not.
IMPORTANCE In 2003, a variant of methicillin-resistant Staphylococcus aureus (MRSA) isolated from pigs was also found in pig farmers in France and the Netherlands. Soon thereafter, this livestock-associated MRSA (LA-MRSA) was identified in many other countries. Transmission of LA-MRSA between humans, particularly in the health care setting, is regarded to occur sporadically. Moreover, studies that describe LA-MRSA transmission used molecular characterization of isolates with limited discriminatory power, making the validity of the conclusion that transmission occurred questionable. In our study, we sequenced the complete genomes of 206 LA-MRSA isolates, obtained from more than 30 presumed LA-MRSA transmission events. Analysis of the data showed that transmission of LA-MRSA between humans had indeed occurred in more than 90% of these events. We conclude that transmission of LA-MRSA between humans does occur in Dutch health care settings; therefore, a decision to discontinue the search and destroy policy for LA-MRSA should be taken with caution.
INTRODUCTION
Methicillin-resistant Staphylococcus aureus (MRSA) is an important cause of hospital-acquired and community-acquired infections (1). In 2003, an MRSA variant cultured from pigs and pig farmers emerged in the Netherlands and France (2, 3). This clonal lineage was designated MLST clonal complex 398 (CC398), and a large number of countries reported CC398 cultured from animal specimens, revealing a worldwide prevalence (4, 5). Besides pigs, CC398 has been found in other livestock animals, such as veal calves and poultry (6, 7), and was therefore designated livestock-associated MRSA (LA-MRSA). In the Netherlands, LA-MRSA CC398 isolated from humans has become the predominant MRSA clade among isolates submitted for typing in the Dutch MRSA surveillance program since 2007 (5, 8).
Despite the high prevalence of LA-MRSA, its transmissibility between humans has been a subject of debate. Transmission of LA-MRSA between animals and from animals to humans has been described in detail, and transmission occurs frequently (9). For human-to-human transmission of LA-MRSA, the capability is less clear, and studies on this subject focused on two topics, namely, transmission between humans in a livestock setting or within a health care setting. Within the livestock setting, transmission of LA-MRSA between humans has been described in livestock farmers, where transmission among broilers, humans, and their environment was found (10). A recent study showed that transmission of LA-MRSA from the livestock veterinarian to household members who reported not having livestock contact occurred in 15 of the 16 investigated households (11). Surveys on the transmission of LA-MRSA within the health care setting showed that LA-MRSA was 4 to 6 times less transmissible than other MRSA lineages (12, 13). In addition, nosocomial transmission of LA-MRSA in Dutch hospitals was reported to be 72% less likely to occur than transmission of non-LA-MRSA (14). On the other hand, outbreaks of LA-MRSA have been described, and two recent reports showed that between 21% and 26%, respectively, of the MRSA isolates cultured from persons without known MRSA risk factors, the so-called MRSA of unknown origin (MUO), were LA-MRSA (15, 16). These MUOs suggest the spread of LA-MRSA through routes other than livestock exposure.
One of the difficulties regarding investigations on transmission routes of LA-MRSA is that this MRSA variant represents a genetically homogenous clade resulting in limited differentiation using frequently used typing techniques, such as multilocus sequence typing (MLST), multilocus variable-number tandem-repeat analysis (MLVA), and Staphylococcus protein A gene (spa) typing (17). Recently, studies using next-generation sequencing (NGS) and whole-genome mapping revealed more genotypic diversity among LA-MRSA and suggested a distinction between livestock- and human-associated CC398 clades (11, 18–20). However, the use of NGS for LA-MRSA in these studies focused mainly on the population structure of LA-MRSA, using isolates from different geographical sources and origins. Studies where NGS was applied for transmission studies have thus far been limited to human-associated non-LA-MRSA clades, such as E-MRSA 15 (21, 22).
In this study, we performed NGS on more than 200 LA-MRSA isolates obtained from humans submitted for Dutch national MRSA surveillance. Together with epidemiological data, we used NGS data to assess the capability of LA-MRSA to be transmitted between humans in Dutch health care settings.
MATERIALS AND METHODS
Bacterial isolates.
Virtually all MRSA isolates (one isolate per person per year) obtained from humans admitted to health care centers are submitted for molecular typing to the National Institute of Public Health and the Environment (RIVM) for Dutch national MRSA surveillance. In addition, medical microbiologists or infection control practitioners fill out questionnaires regarding epidemiologic risk factors for MRSA colonization or infection, including contact with livestock, for the persons from whom MRSA was cultured. In this study, LA-MRSA was defined as isolates with MLVA types belonging to MLVA complex 398 (MC398).
To assess the capability of LA-MRSA to cause nosocomial transmission, we included 12 LA-MRSA isolates from a presumed outbreak in a Dutch health care facility and 12 isolates from an outbreak in a Dutch nursing home (23). Furthermore, inspection of the 9,698 questionnaires obtained from 2008 to 2012 showed that in 1,291 cases, MRSA was isolated because nosocomial transmission was suspected. In 673 of these presumed transmission events, isolates were sent to the RIVM for typing. Of those, 41 involved LA-MRSA, but in 15 events, the isolates of the index and secondary cases did not have the same MLVA/spa types. As a result, 26 presumed LA-MRSA nosocomial transmission events comprising 60 isolates were included in this study. Besides these 26 events, 4 presumed transmission events, comprising 8 isolates, in Dutch health care settings, as described by van Rijen et al. (16), were also included, resulting in a total of 30 investigated events. Of the 30 presumptive transmission events, 25 were single transmissions, and the other 5 involved multiple transmissions.
In addition to these presumed LA-MRSA transmission isolates, 114 LA-MRSA isolates comprising the 3 predominant LA-MRSA MLVA/spa types (MT398/t011, MT572/t108, and MT569/t034) in the Netherlands obtained between 2003 and 2012 were included for analysis to provide epidemiological context. All available MT398/t011, MT572/t108, and MT569/t034 isolates from 2003 (n = 12), 2004 (n = 12), and 2005 (n = 10) and the first 5 isolates (if available) of those 3 types from 2006 until 2012 were used.
All 206 isolates were previously characterized using spa typing, MLVA, and whole-genome mapping (17, 24, 25).
Next-generation sequencing.
All 206 LA-MRSA isolates included in this study were subjected to NGS. In total, 148 LA-MRSA isolates, originating from the 3 predominant LA-MRSA MLVA/spa types, the previously described outbreak (23), the presumed LA-MRSA outbreak, and the 5 presumed nosocomial transmission events (16), were sequenced as part of the 100K genome project by University of California—Davis on a HiSeq 2000 (http://100kgenome.vetmed.ucdavis.edu/). The other 58 isolates were commercially sequenced on a HiSeq 2500 sequencer (BaseClear, Leiden, Netherlands).
Core genome single nucleotide variant (SNV) analysis.
The complete, annotated genome of LA-MRSA strain RIVM1295 with MT572/t108 (accession number CP013616), isolated from a Dutch patient was determined by NGS. Furthermore, complete, annotated chromosomes from the two other LA-MRSA isolates obtained from Dutch patients representing the two other dominant MLVA/spa types, MT398/t011 (RIVM1607, CP013619) and MT569/t034 (RIVM3897, CP013621), were used for comparison with the reference genome RIVM1295 to exclude noncore genome regions in the SNV analysis (see Fig. S1 in the supplemental material). In total, 30 regions, mostly comprising genes encoding transposases (n = 10), a single bacteriophage, small regions flanking the insertions sites of bacteriophages present in strains RIVM1607 and RIVM3897 (n = 8), and the 6 rRNA gene regions, were excluded. The regions that were excluded from the core genome are indicated as annotations in the complete genome sequence (CP013616). The CLC bio Genomics server/workbench version 7.5 (CLC bio, Aarhus, Denmark) was used for the identification of SNVs, and SNV data were imported into Bionumerics version 7.5 for comparative analyses (Applied Maths, Sint-Martens-Latem, Belgium).
Staphylococcal cassette chromosome mec and plasmid identification in LA-MRSA isolates.
We assessed the staphylococcal cassette chromosome mec (SCCmec) region and plasmid composition of LA-MRSA isolates to collect additional information from the accessory genome. The SCCmec region of all 206 LA-MRSA isolates was mapped against all known SCCmec references (www.sccmec.org) available in the NCBI database in October 2015, and SCCmec types were assigned if a complete match was found. Divergent variants were named after the most similar SCCmec type followed by an extension to indicate their distinctive characteristics.
To study the diversity of plasmids among Dutch LA-MRSA isolates, we used the sequences of three plasmids (accession numbers AM990993, AM990994, and AM990995) found in ST398 reference strain S0385 (accession number AM990992) and sequences obtained from the NCBI database of six plasmids isolated from ST398 strains and sequenced by other researchers (Table 1) (27–33). Furthermore, sequences from three plasmids, pRIVM1295-1 (CP013617), pRIVM1295-2 (CP013618), and pRIVM1607 (CP013620), identified in our in-house LA-MRSA reference strains were also used. During screening of the 206 LA-MRSA isolates against the plasmids, NGS reads of 53 LA-MRSA isolates mapped against only parts of the plasmids pS0385-2, pS0385-3, and pKKS627. De novo assembly and a blast of the rep genes against the NCBI database revealed the complete sequence of 5 novel plasmids and 3 novel incomplete plasmids in the collection, resulting in 20 plasmids used for screening.
TABLE 1.
Overview of plasmid screening among 206 LA-MRSA isolates
| No. | Plasmid | Prevalence (no.) | Resistance gene(s) | Accession no. | Size (kb) | Reference |
|---|---|---|---|---|---|---|
| 1 | pRIVM1295-1 | 61 | ermC | CP013617 | 2.4 | This study |
| 2 | pRIVM4294 | 14 | CP013625 | 3.1 | This study | |
| 3 | pRIVM1295-2 | 13 | CP013618 | 3.0 | This study | |
| 4 | pRIVM4296 | 13 | CP013626 | 3.0 | This study | |
| 5 | pRIVM1183 | 9 | tetL, dfrK, ermT | CP013627 | 7.3a | This study |
| 6 | pRIVM4390 | 7 | aadD | CP013623 | 4.6 | This study |
| 7 | pRIVM0677 | 5 | CP013622 | 4.4 | This study | |
| 8 | pRIVM1076 | 3 | CP013624 | 2.2 | This study | |
| 9 | pRIVM1607 | 1 | CP013620 | 1.5 | This study | |
| 10 | pRIVM4293 | 1 | tetL, dfrK, aadD | CP013628 | 6.2a | This study |
| 11 | pRIVM4256 | 1 | tetL, dfrK, aadD | CP013629 | 6.7a | This study |
| 12 | pS0385-1 | 104 | tetK | AM990993 | 5.2 | 26 |
| 13 | pS0385-2 | 11 | aadD | AM990994 | 4.4 | 26 |
| 14 | pS0385-3 | 2 | AM990995 | 3.2 | 26 | |
| 15 | pSWS2889 | 3 | spd | HG803547 | 3.9 | 27 |
| 16 | pKKS627 | 2 | tetL, dfrK | FN390948 | 6.2 | 28 |
| 17 | pKKS825 | 0 | tetL, dfrK, vgaC, aadD | NC_013034 | 14.3 | 29 |
| 18 | pKKS966 | 0 | dfrK | FN677368 | 5.0 | 30 |
| 19 | pUR2940 | 0 | tetL, dfrK, ermC, ermT | NG_041022 | 20 | 25 |
| 20 | pNVH01 | 0 | qacJ | AJ512814 | 2.7 | 24 |
Plasmid with incomplete sequence.
Accession number(s).
The novel plasmid sequences were submitted to the NCBI database and are available under accession numbers CP013622 to CP013629.
RESULTS
SNV analysis.
In total, 6,461 single nucleotide variants (SNVs) were identified among the 206 isolates in the collection and used for comparison (Table 2; see also Fig. S1 in the supplemental material). Most SNVs were found in coding regions of the chromosome and comprised nonsilent (n = 2,920), silent (n = 1,768), and missense mutations (n = 104), while 1,669 SNVs were identified in the noncoding regions. The majority of SNVs represented transitions (n = 4,467; 69%), whereas 1,994 SNVs were transversions.
TABLE 2.
Single nucleotide variants among the 206 LA-MRSA isolatesa
| Mutation | SNV | ncSNV |
sSNV |
nsSNV |
msSNV |
||||
|---|---|---|---|---|---|---|---|---|---|
| No. | % | No. | % | No. | % | No. | % | ||
| Transition | A↔G | 521 | 31.2 | 631 | 35.7 | 958 | 32.8 | 31 | 29.8 |
| C↔T | 570 | 34.2 | 743 | 42.0 | 983 | 33.7 | 30 | 28.8 | |
| Transversion | A↔T | 200 | 12.0 | 182 | 10.3 | 251 | 8.6 | 18 | 17.3 |
| A↔C | 145 | 8.7 | 96 | 5.4 | 299 | 10.2 | 10 | 9.6 | |
| G↔T | 174 | 10.4 | 92 | 5.2 | 291 | 10.0 | 15 | 14.4 | |
| G↔C | 59 | 3.5 | 24 | 1.4 | 138 | 4.7 | 0 | 0.0 | |
| All SNVs | 1,669 | 1,768 | 2,920 | 104 | |||||
nc, noncoding; s, silent; ns, nonsilent; ms, missense.
The minimum spanning tree based on SNV analysis showed that the 206 isolates used in this study mainly clustered into two groups. One group contained virtually all MT398/t011 isolates and two isolates with MT566/t1456 (Fig. 1). The average SNV distance between members of the MT398/t011 group was 31 SNVs, with a range of 3 to 115 SNVs between two isolates in this group. The other group contained all MT572/t108 isolates that clustered closely together, with an average distance of 39 SNVs and a range of 6 to 85 SNVs between two MT572/t108 isolates. There was a distance of 265 SNVs between the closest members of the MT572/t108 and MT398/t011 isolates. The MT569/t034 isolates were genetically more diverse and did not cluster in a group. The range of SNVs between two MT569/t034 isolates was 3 to 398 SNVs.
FIG 1.
Minimum spanning tree based on SNV analysis of 206 LA-MRSA isolates. The tree was based on 6,461 SNV positions, and clustering was done using a categorical coefficient. Each isolate in the tree is displayed as a circle. The colors represent the MLVA/spa types, and the lines between the isolates denote the distance in number of SNVs.
SCCmec analysis.
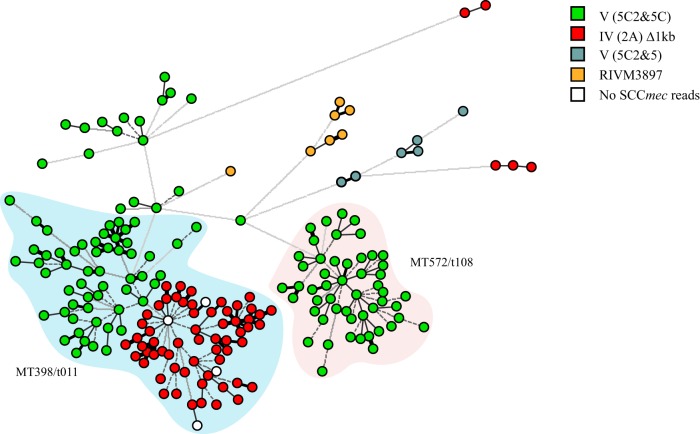
In total, 4 different SCCmec types could be assigned in 202 of the 206 LA-MRSA isolates. Within the 4 SCCmec types, 16 different SCCmec variants with distinct compositions were identified (see Fig. S2 in the supplemental material). Most isolates (n = 99) carried SCCmec type V (5C2&5C). (The structural type is indicated by a Roman numeral, with a lowercase letter indicating the subtype, and the ccr complex and the mec complex are indicated by an Arabic numeral and an uppercase letter, respectively. Where there is an extra ccr element, this is indicated by “&” and an Arabic numeral designating the ccr type.) A variant of type V (5C2&5C), from which a 5.2-kb fragment comprising 2 transposase-encoding genes and the tetK gene were deleted, was found in 15 isolates. Six other variants of type V (5C2&5C), with deletions that ranged from 3 kb to 22 kb, were present in 11 isolates. The second most frequently identified SCCmec type was type IV (2A) and was carried by 59 isolates. However, all isolates had a 1-kb deletion in the distal part of the SCCmec region compared to the SCCmec reference. Five isolates carried variants of SCCmec type IV (2A), with deletions of 1.2 kb (n = 3) or 2.2 kb (n = 2). A third SCCmec type, present in 7 isolates, did not match any known reference and was designated SCCmec type RIVM3897. A single isolate carried the complete cassette, while the remaining six isolates had a 1-kb deletion. The fourth SCCmec type was V (5C2&5). Two isolates carried a variant with 1.2-kb and 1.6-kb deletions, and four isolates carried 1-kb, 1.2-kb, and 1.6-kb deletions. The NGS data of four isolates contained no reads representing the SCCmec region. Consequently, no SCCmec type could be assigned to the four remaining isolates. Plotting of the four SCCmec types in a minimum spanning tree based on SNVs revealed that type V (5C2&5C) was present in all MT572/t108 isolates, while in isolates yielding MT398/t011, a clear distinction was observed between type V (5C2&5C) and type IV (2A) isolates (Fig. 2). These two SCCmec subgroups did not differ in the average number of SNVs (34 versus 30) or SNV range (1 to 115 versus 2 to 72).
FIG 2.
Distribution of the SCCmec types among 206 LA-MRSA isolates. The tree was based on 6,461 SNV positions, and clustering was done using a categorical coefficient. Each isolate in the tree is displayed as a circle. The colors represent the different SCCmec types. LA-MRSA isolates for which no NGS reads for the SCCmec region were present are indicated as white circles. The blue halo contains all isolates with MLVA/spa type MT398/t011, and the red halo contains all MT572/t108 isolates.
Plasmid screening.
Based on screening for the presence of 20 plasmids, we identified 16 different plasmids among the 206 LA-MRSA isolates. The predominant plasmids were pS0385-1 (n = 104), pRIVM1295-1 (n = 61), and pRIVM4294 (n = 14), while plasmids pNVH01, pKKS825, pKKS966, and pUR2940 were not present (Table 2). The 16 plasmids identified in isolates carried a variety of resistance genes, but tetL (tetracycline), dfrK (trimethoprim), and aadD (kanamycin and neomycin) were predominant.
Plotting of the two predominant plasmids pS0385-1 and pRIVM1295-1 in the minimum spanning tree based on the SNV analysis showed that there was no clear relationship between SNV branches and the presence of plasmid pRIVM1295-1 (see Fig. S5A in the supplemental material). In contrast, plasmid pS0385-1 was only present in isolates carrying SCCmec type V (5C2&5C) (see Fig. S5B). Analysis showed that, in the reference sequence of SCCmec type (KF593809), the 5.2-kb plasmid sequence of pS0385-1 was an integral part of the SCCmec region and was flanked by two transposons. This suggested that the 5.2-kb region might have been misclassified as plasmid pS0385-1. This was corroborated by comparison of the whole-genome map of strain S0385 and its in silico counterpart based on the whole-genome sequence (AM990992) (see Fig. S5C). The in silico map of S0385 lacked a 5-kb segment that was present in the real map of S0385. Strain RIVM5890, for which no NGS reads mapping with the 5-kb segment were found, also lacked this segment in the whole-genome map.
NGS of 2 LA-MRSA outbreaks.
Isolates originating from 2 different outbreaks were subjected to NGS analysis. The first set of 12 isolates belonged to a previously reported outbreak in a Dutch nursing home (23). SNV analysis of the isolates showed a maximum of 23 SNVs between the isolate from the index patient and other outbreak isolates (Fig. 3). The SCCmec region of these isolates was type V (5C2&5C), with a 5.2-kb deletion that included the tetK gene. This SCCmec variant was only found among these 12 LA-MRSA isolates and in 3 other nonrelated isolates. None of the 20 plasmids were present in the isolates of this outbreak.
FIG 3.
SNV analysis of two LA-MRSA outbreaks displaying clustering indicative for transmission. The tree was based on 6,461 SNV positions, and clustering was done using a categorical coefficient. Each node in this minimum spanning tree represents a single LA-MRSA isolate. Nodes with identical colors represent isolates from the same LA-MRSA outbreak. On the right side, the SNV positions relative to the sequence of the isolate of the index patients of each of the outbreaks are given. Each colored ring represents a single isolate belonging to the outbreak.
The second LA-MRSA outbreak, also comprising 12 isolates, occurred in a Dutch hospital. The number of SNVs between the outbreak isolates and the first isolate from the index patient ranged from 17 to 25 SNVs. The second isolate of the index patient, taken from the toe, differed in 59 SNVs from the first isolate of the index patient, which originated from a throat/nose/perineum sample. All isolates carried SCCmec type IV (2A). All isolates carried plasmid pRIVM1295-1 except for the second isolate of the index patient. The second isolate of the index patient carried another plasmid, designated pRIVM4390, that was also found in one of the other outbreak isolates but was lacking from the other isolates. Although a subset of SNVs was present in multiple isolates in both outbreaks, the transmission route could not be inferred from the distribution of accumulated SNVs.
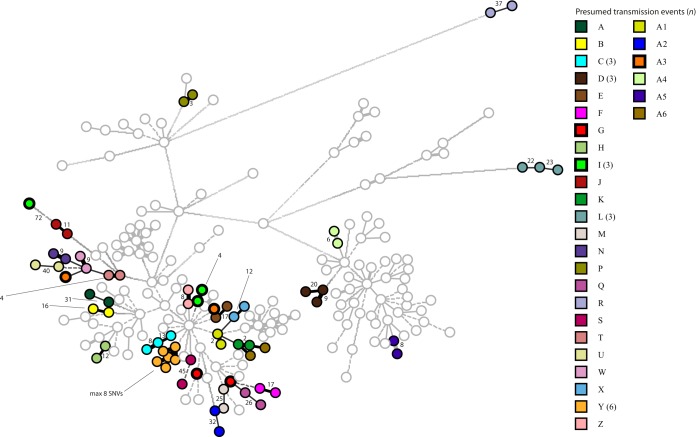
NGS of the 30 presumed LA-MRSA transmission events.
In the 25 presumed single transmission events, a difference ranging from 2 to 105 SNVs between the isolates of a pair belonging to a transmission was found (Table 3; Fig. 4). In 22 of the 25 cases, the pairs of isolates carried the same SCCmec type. The exceptions were two isolates of two presumed transmissions that carried no SCCmec, while in a single event (A3), different SCCmec types were carried by the isolates, namely, types V (5C2&5) and IV (2A)Δ1kb. In 11 of these 25 transmissions, the pairs of isolates also carried the same plasmids. None of the 20 plasmids were present in both isolates of 8 other pairs, while different plasmids were found in the isolates of the remaining 6 pairs. In 5 of the 6 pairs, the plasmids present in the isolate of the index patient were absent in the isolate of the contact. In the other pair, the isolate of the contact carried plasmid pRIVM1295-2, which was absent in the isolate of the index patient.
TABLE 3.
NGS of the 25 presumed single LA-MRSA transmission events
| Event | MLVA/spa | No. of SNVs | SCCmec | Shared plasmid(s) | Differentiating plasmid(s) |
|---|---|---|---|---|---|
| A1 | MT398/t011 | 2 | IV (2A)Δ1kb | pRIVM1295-1, pRIVM4390 | |
| K | MT398/t011 | 2 | IV (2A)Δ1kb | pRIVM4390 | |
| P | MT569/t034 | 3 | V (5C2&5C) | pRIVM0677, pRIVM4296 | |
| T | MT398/t011 | 4 | V (5C2&5C) | pS0385-2 | |
| A4 | MT572/t108 | 6 | V (5C2&5C) | pS0385-2a | |
| A5 | MT572/t108 | 8 | V (5C2&5C) | pRIVM1076 | |
| Z | MT398/t011 | 8 | IV (2A)Δ1kb | pRIVM1183 | |
| N | MT398/t011 | 9 | V (5C2&5C) | ||
| W | MT398/t011 | 9 | V (5C2&5C) | pS0385-2 | |
| J | MT398/t011 | 11 | V (5C2&5C) | pRIVM1295-1, pRIVM1295-2, pKKS627 | |
| H | MT398/t011 | 12 | V (5C2&5C) | ||
| X | MT398/t011 | 12 | IV (2A)Δ1kb | pRIVM1295-1a, pRIVM4390a | |
| B | MT398/t011 | 16 | V (5C2&5C) | pRIVM1295-2 | |
| E | MT398/t011 | 17 | IV (2A)Δ1kb, no SCCmec | pRIVM1295-1 | |
| F | MT398/t011 | 17 | IV (2A)Δ1kb | ||
| A6 | MT398/t011 | 19 | IV (2A)Δ1kb | ||
| M | MT398/t011 | 25 | IV (2A)Δ1kb | pRIVM1183 | |
| Q | MT398/t011 | 26 | IV (2A)Δ1kb | pRIVM4294 | |
| A | MT398/t011 | 31 | V (5C2&5C) | ||
| A2 | MT398/t011 | 32 | IV (2A)Δ1kb, no SCCmec | pRIVM1295-1a, pRIVM1076a | |
| R | MT567/t899 | 37 | IV (2A)Δ1kb & Δ2kb | ||
| U | MT398/t011 | 40 | V (5C2&5C) | ||
| S | MT566/t1456 | 45 | IV (2A)Δ1kb | ||
| G | MT398/t011 | 51 | IV (2A)Δ1kb | pRIVM1295-1a | |
| A3 | MT398/t011 | 105 | V (5C2&5C), IV (2A)Δ1kb | pRIVM4256a |
Plasmid present in the isolate of the index case only.
FIG 4.
Minimum spanning tree displaying clustering indicative for transmission of LA-MRSA between index patients and contacts in Dutch health care settings. The tree was based on 6,461 SNV positions, and clustering was done using a categorical coefficient. Each node in this minimum spanning tree represents a single LA-MRSA isolate. Nodes with identical colors represent isolates from the same presumed transmission event. Circles with thicker lines represent isolates where transmission seems unlikely. The number of SNVs between isolates of the same presumed transmission is indicated adjacent to the line connecting the nodes in the minimum spanning tree.
In 4 of the 5 cases (C, D, I, L, and Y), the isolates associated with the same multiple presumed nosocomial transmission event clustered closely together (Table 4; Fig. 4). The exception was event I, where 126 SNVs between the isolate of the index and the isolate of one of the two contacts were found. The isolate of the other contact differed only in 4 SNVs from the isolate of the index patient. In 4 of the 5 events, the same SCCmec type was carried by all isolates. In the remaining event (I, n = 3 isolates), SCCmec type IV (2A) was present in 2 isolates that differed in only 4 SNVs, but the other isolate differing in 126 SNVs carried an SCCmec type V (5C2&5C). An identical plasmid composition was found in the isolates of event Y, where pRIVM1295-1 was carried by all isolates. Different plasmid compositions between the isolates were seen in events C, D, and I, while no plasmids were found in isolates from event L. In event C, the isolate of the index patient carried pRIVM1295-1, and this plasmid was also present in one of the contacts, but the isolate of this person also acquired plasmid pRIVM1295-2. In event D, the isolates of the index patient and one of the contacts carried plasmid pRIVM1295-1, while no plasmids were present in the isolate of the other contact. In the remaining event (I), plasmid pRIVM1295-2 was found in the isolates that differed in 4 SNVs. The plasmid pRIVM1295-2 was absent from the isolate of the contact that differed in 126 SNVs, but this isolate carried plasmid pRIVM1295-1 instead.
TABLE 4.
NGS of the multiple presumed nosocomial LA-MRSA transmission events
| Event group | Event | MLVA/spa | No. of SNVs | SCCmec | Plasmid(s) |
|---|---|---|---|---|---|
| Y | Index | MT398/t011 | IV (2A)Δ1kb | pRIVM1295-1 | |
| Event 1 | MT398/t011 | 5 | IV (2A)Δ1kb | pRIVM1295-1 | |
| Event 2 | MT398/t011 | 8 | IV (2A)Δ1kb | pRIVM1295-1 | |
| Event 3 | MT398/t011 | 5 | IV (2A)Δ1kb | pRIVM1295-1 | |
| Event 4 | MT398/t011 | 7 | IV (2A)Δ1kb | pRIVM1295-1 | |
| Event 5 | MT398/t011 | 3 | IV (2A)Δ1kb | pRIVM1295-1 | |
| C | Index | MT398/t011 | IV (2A)Δ1kb | pRIVM1295-1 | |
| Event 1 | MT398/t011 | 8 | IV (2A)Δ1kb | No plasmids | |
| Event 2 | MT398/t011 | 13 | IV (2A)Δ1kb | pRIVM1295-1, pRIVM1295-2 | |
| D | Index | MT572/t108 | V (5C2&5C) | pRIVM1295-1 | |
| Event 1 | MT572/t108 | 9 | V (5C2&5C) | No plasmids | |
| Event 2 | MT572/t108 | 20 | V (5C2&5C) | pRIVM1295-1 | |
| L | Index | MT569/t034 | IV (2A)Δ1kb & Δ1.2kb | No plasmids | |
| Event 1 | MT569/t034 | 27 | IV (2A)Δ1kb & Δ1.2kb | No plasmids | |
| Event 2 | MT569/t034 | 23 | IV (2A)Δ1kb & Δ1.2kb | No plasmids | |
| I | Index | MT398/t011 | IV (2A)Δ1kb | pRIVM1295-2 | |
| Event 1 | MT398/t011 | 126 | V (5C2&5C) | pRIVM1295-1 | |
| Event 2 | MT398/t011 | 4 | IV (2A)Δ1kb | pRIVM1295-2 |
In the NGS data of two presumed single transmission events, no reads for the SCCmec were found in one of the isolates in each of two events (A2 and E). However, the number of SNVs between the isolates belonging to the same pair was low, amounting to 32 and 17 SNVs, respectively. In addition, whole-genome mapping, previously performed on a different DNA sample of the isolates, showed indistinguishable fragments in the SCCmec region, indicating that the SCCmec was previously present in both isolates. Repeated PCR analysis of the DNA preparations used for NGS revealed that this batch of DNA of the two aberrant isolates did not contain a mec gene, corroborating the NGS analysis.
DISCUSSION
In this study, we used NGS data of 206 LA-MRSA isolates obtained from humans and found that nosocomial transmission of LA-MRSA in Dutch health care facilities does occur. Transmissions were inferred from SNV data, and conclusions were supported by data on the composition of the SCCmec region. However, variation in the SCCmec region is too limited to use only this region as a genetic marker in transmission studies. This is also true for the use of data on the presence and composition of plasmids. In addition, plasmids appear to be lost and acquired quite rapidly after LA-MRSA is transmitted from one patient to the other, making plasmid presence a poor marker to assess whether transmission has occurred.
In total, 32 LA-MRSA presumed transmission events comprising 25 single events, 5 multiple events, and 2 outbreaks, each comprising 12 patients, were studied. In the majority of the putative transmission events, the isolates of the index patient and the contact(s) clustered closely together in an SNV-based minimum spanning tree with 2 to 45 SNVs between the isolates. In only 4 presumed transmission events did the number of SNVs between isolates of the index and the contact exceed 50. In 3 of those events, the isolates did not cluster together, indicating that transmission was unlikely. In the remaining event, 1 of 2 isolates, obtained from different anatomical locations of the presumed index of an outbreak, differed by 59 SNVs from its closest related outbreak isolate. However, it obviously belonged to the outbreak, suggesting that an LA-MRSA strain colonizing or infecting different sites within the same person may evolve independently over time. Recent reports that applied whole-genome sequencing on multiple colonies from the same person also identified a cloud of diversity among the isolates (21, 34, 35). These studies were carried out on ST22 and ST239 MRSA isolates, but similar findings might be expected from LA-MRSA.
This study showed that the number of SNVs alone is not sufficient to assess whether transmission has occurred. Combining NGS data and epidemiological data is essential to determine transmission, and the use of SNV data of epidemiologically unrelated isolates as context proved extremely helpful. Furthermore, analysis of NGS data other than SNVs may be used to provide additional information to assess transmission events. Although variation of the SCCmec region turned out to be limited in LA-MRSA, it provided supportive evidence for several transmission events. For example, all isolates from a previously reported LA-MRSA outbreak had a distinctive 5.2-kb deletion in the SCCmec region that supported the conclusion that transmission was indeed likely. Conversely, differences in SCCmec types provided further proof that transmission did not occur in other cases. Unexpectedly, two isolates belonging to two different transmission events and previously identified as LA-MRSA carried no SCCmec cassette. The most likely explanation for this observation is that the isolates consisted of heterogenetic populations comprising both SCCmec-positive and -negative variants and that the SCCmec-positive variants were lost during subculturing to prepare DNA for NGS.
The plasmid composition was identical in isolates of some of the transmissions. However, no plasmids were found in other events, and in several cases, differences in plasmid composition were observed, although SNV and SCCmec analysis indicated that transmission was likely. Stanczak-Mrozek et al. recently showed that MRSA variants that have acquired or lost mobile genetic elements were common in nasally colonized populations (36). Furthermore, a study using CC398 isolates showed that horizontal gene transfer, including plasmids, occurred at a very high frequency in vivo (26). Since only a single colony was used for the initial culture from the clinical material and for various subcultures, different variants of the same strain, with or without plasmids, may have been sequenced, resulting in the observed differences.
Our study has a number of limitations. First, all sequenced isolates belonged to the LA-MRSA (MC398) clade. This limits a comparison of NGS data between LA-MRSA and other MRSA variants and hampers studies on the transmission rates of the different MRSA clades in Dutch health care facilities. Second, the presumed nosocomial transmission events were selected based on the epidemiological data provided by the medical microbiology laboratories and affiliated infection prevention practitioners. We do not know whether all transmission events were correctly identified, and this could lead to either under- or over-representation of the number of presumed nosocomial transmissions. Finally, our NGS data were only screened for plasmids that were associated with livestock. Other plasmids were not taken into account, and this could have provided more information. Furthermore, the use of data from other mobile genetic elements, such as bacteriophages, was not included in this study (36). In conclusion, our study strongly suggests that transmission of LA-MRSA in Dutch health care settings does occur. NGS could confirm previously reported transmission events and indicated that transmission was unlikely in three presumed transmissions. We conclude that investigations regarding transmissions of LA-MRSA should be supported by epidemiological data and be investigated using the context of epidemiologically related and unrelated isolates. Analysis of the SCCmec region provided useful information to support SNV analysis, but plasmid identification did not.
Supplementary Material
ACKNOWLEDGMENTS
We thank Rebecca te Riet from Infection Control, Ziekenhuisgroep Twente, Almelo, the Netherlands, for supplying the isolates of the presumed hospital outbreak. Furthermore, we thank all medical microbiology laboratories for sending their MRSA isolates to the RIVM. Finally, we thank the infection prevention practitioners for filling in and supplying the questionnaires.
The funding source had no role in the study design, data collection, data analysis, data interpretation, or writing of the report. The corresponding author had access to all the data of the study and the final responsibility to submit for publication.
Funding Statement
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sector.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/AEM.00773-16.
REFERENCES
- 1.Vandenesch F, Naimi T, Enright MC, Lina G, Nimmo GR, Heffernan H, Liassine N, Bes M, Greenland T, Reverdy ME, Etienne J. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg Infect Dis 9:978–984. doi: 10.3201/eid0908.030089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armand-Lefevre L, Ruimy R, Andremont A. 2005. Clonal comparison of Staphylococcus aureus isolates from healthy pig farmers, human controls, and pigs. Emerg Infect Dis 11:711–714. doi: 10.3201/eid1105.040866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Voss A, Loeffen F, Bakker J, Klaassen C, Wulf M. 2005. Methicillin-resistant Staphylococcus aureus in pig farming. Emerg Infect Dis 11:1965–1966. doi: 10.3201/eid1112.050428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhat M, Dumortier C, Taylor BS, Miller M, Vasquez G, Yunen J, Brudney K, Sanchez EJ, Rodriguez-Taveras C, Rojas R, Leon P, Lowy FD. 2009. Staphylococcus aureus ST398, New York City and Dominican Republic. Emerg Infect Dis 15:285–287. doi: 10.3201/eid1502.080609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Cleef BA, Monnet DL, Voss A, Krziwanek K, Allerberger F, Struelens M, Zemlickova H, Skov RL, Vuopio-Varkila J, Cuny C, Friedrich AW, Spiliopoulou I, Paszti J, Hardardottir H, Rossney A, Pan A, Pantosti A, Borg M, Grundmann H, Mueller-Premru M, Olsson-Liljequist B, Widmer A, Harbarth S, Schweiger A, Unal S, Kluytmans JA. 2011. Livestock-associated methicillin-resistant Staphylococcus aureus in humans, Europe. Emerg Infect Dis 17:502–505. doi: 10.3201/eid1703.101036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Graveland H, Wagenaar JA, Verstappen KM, Oosting-van Schothorst I, Heederik DJ, Bos ME. 2012. Dynamics of MRSA carriage in veal calves: a longitudinal field study. Prev Vet Med 107:180–186. doi: 10.1016/j.prevetmed.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 7.Nemati M, Hermans K, Lipinska U, Denis O, Deplano A, Struelens M, Devriese LA, Pasmans F, Haesebrouck F. 2008. Antimicrobial resistance of old and recent Staphylococcus aureus isolates from poultry: first detection of livestock-associated methicillin-resistant strain ST398. Antimicrob Agents Chemother 52:3817–3819. doi: 10.1128/AAC.00613-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huijsdens XW, Bosch T, van Santen-Verheuvel MG, Spalburg E, Pluister GN, van Luit M, Heck ME, Haenen A, de Neeling AJ. 2009. Molecular characterisation of PFGE nontypable methicillin-resistant Staphylococcus aureus in the Netherlands, 2007. Euro Surveill 14:pii=19335 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleID=19335. [DOI] [PubMed]
- 9.Crombé F, Vanderhaeghen W, Dewulf J, Hermans K, Haesebrouck F, Butaye P. 2012. Colonization and transmission of methicillin-resistant Staphylococcus aureus ST398 in nursery piglets. Appl Environ Microbiol 78:1631–1634. doi: 10.1128/AEM.07356-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wendlandt S, Kadlec K, Fessler AT, Mevius D, van Essen-Zandbergen A, Hengeveld PD, Bosch T, Schouls L, Schwarz S, van Duijkeren E. 2013. Transmission of methicillin-resistant Staphylococcus aureus isolates on broiler farms. Vet Microbiol 167:632–637. doi: 10.1016/j.vetmic.2013.09.019. [DOI] [PubMed] [Google Scholar]
- 11.Bosch T, Verkade E, van Luit M, Landman F, Kluytmans J, Schouls LM. 2015. Transmission and persistence of livestock-associated MRSA among veterinarians and their household members. Appl Environ Microbiol 81:124–129. doi: 10.1128/AEM.02803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bootsma MC, Wassenberg MW, Trapman P, Bonten MJ. 2011. The nosocomial transmission rate of animal-associated ST398 meticillin-resistant Staphylococcus aureus. J R Soc Interface 8:578–584. doi: 10.1098/rsif.2010.0349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hetem DJ, Bootsma MC, Troelstra A, Bonten MJ. 2013. Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus. Emerg Infect Dis 19:1797–1802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wassenberg MW, Bootsma MC, Troelstra A, Kluytmans JA, Bonten MJ. 2011. Transmissibility of livestock-associated methicillin-resistant Staphylococcus aureus (ST398) in Dutch hospitals. Clin Microbiol Infect 17:316–319. doi: 10.1111/j.1469-0691.2010.03260.x. [DOI] [PubMed] [Google Scholar]
- 15.Lekkerkerk WS, van de Sande-Bruinsma N, van der Sande MA, Tjon ATA, Groenheide A, Haenen A, Timen A, van den Broek PJ, van Wamel WJ, de Neeling AJ, Richardus JH, Verbrugh HA, Vos MC. 2012. Emergence of MRSA of unknown origin in the Netherlands. Clin Microbiol Infect 18:656–661. doi: 10.1111/j.1469-0691.2011.03662.x. [DOI] [PubMed] [Google Scholar]
- 16.van Rijen MM, Bosch T, Verkade EJ, Schouls L, Kluytmans JA, CAM Study Group. 2014. Livestock-associated MRSA carriage in patients without direct contact with livestock. PLoS One 9:e100294. doi: 10.1371/journal.pone.0100294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bosch T, Verkade E, van Luit M, Pot B, Vauterin P, Burggrave R, Savelkoul P, Kluytmans J, Schouls L. 2013. High-resolution typing by whole genome mapping enables discrimination of LA-MRSA (CC398) strains and identification of transmission events. PLoS One 8:e66493. doi: 10.1371/journal.pone.0066493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Price LB, Stegger M, Hasman H, Aziz M, Larsen J, Andersen PS, Pearson T, Waters AE, Foster JT, Schupp J, Gillece J, Driebe E, Liu CM, Springer B, Zdovc I, Battisti A, Franco A, Zmudzki J, Schwarz S, Butaye P, Jouy E, Pomba C, Porrero MC, Ruimy R, Smith TC, Robinson DA, Weese JS, Arriola CS, Yu F, Laurent F, Keim P, Skov R, Aarestrup FM. 2012. Staphylococcus aureus CC398: host adaptation and emergence of methicillin resistance in livestock. mBio 3:e00305-11. doi: 10.1128/mBio.00305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stegger M, Liu CM, Larsen J, Soldanova K, Aziz M, Contente-Cuomo T, Petersen A, Vandendriessche S, Jimenez JN, Mammina C, van Belkum A, Salmenlinna S, Laurent F, Skov RL, Larsen AR, Andersen PS, Price LB. 2013. Rapid differentiation between livestock-associated and livestock-independent Staphylococcus aureus CC398 clades. PLoS One 8:e79645. doi: 10.1371/journal.pone.0079645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Uhlemann AC, Porcella SF, Trivedi S, Sullivan SB, Hafer C, Kennedy AD, Barbian KD, McCarthy AJ, Street C, Hirschberg DL, Lipkin WI, Lindsay JA, DeLeo FR, Lowy FD. 2012. Identification of a highly transmissible animal-independent Staphylococcus aureus ST398 clone with distinct genomic and cell adhesion properties. mBio 3:e00027-12. doi: 10.1128/mBio.00027-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Harris SR, Cartwright EJ, Torok ME, Holden MT, Brown NM, Ogilvy-Stuart AL, Ellington MJ, Quail MA, Bentley SD, Parkhill J, Peacock SJ. 2013. Whole-genome sequencing for analysis of an outbreak of methicillin-resistant Staphylococcus aureus: a descriptive study. Lancet Infect Dis 13:130–136. doi: 10.1016/S1473-3099(12)70268-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koser CU, Holden MT, Ellington MJ, Cartwright EJ, Brown NM, Ogilvy-Stuart AL, Hsu LY, Chewapreecha C, Croucher NJ, Harris SR, Sanders M, Enright MC, Dougan G, Bentley SD, Parkhill J, Fraser LJ, Betley JR, Schulz-Trieglaff OB, Smith GP, Peacock SJ. 2012. Rapid whole-genome sequencing for investigation of a neonatal MRSA outbreak. N Engl J Med 366:2267–2275. doi: 10.1056/NEJMoa1109910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Verkade E, Bosch T, Hendriks Y, Kluytmans J. 2012. Outbreak of methicillin-resistant Staphylococcus aureus ST398 in a Dutch nursing home. Infect Control Hosp Epidemiol 33:624–626. doi: 10.1086/665726. [DOI] [PubMed] [Google Scholar]
- 24.Harmsen D, Claus H, Witte W, Rothganger J, Claus H, Turnwald D, Vogel U. 2003. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J Clin Microbiol 41:5442–5448. doi: 10.1128/JCM.41.12.5442-5448.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schouls LM, Spalburg EC, van Luit M, Huijsdens XW, Pluister GN, van Santen-Verheuvel MG, van der Heide HG, Grundmann H, Heck ME, de Neeling AJ. 2009. Multiple-locus variable number tandem repeat analysis of Staphylococcus aureus: comparison with pulsed-field gel electrophoresis and spa typing. PLoS One 4:e5082. doi: 10.1371/journal.pone.0005082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy AJ, Loeffler A, Witney AA, Gould KA, Lloyd DH, Lindsay JA. 2014. Extensive horizontal gene transfer during Staphylococcus aureus co-colonization in vivo. Genome Biol Evol 6:2697–2708. doi: 10.1093/gbe/evu214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kadlec K, Schwarz S. 2009. Novel ABC transporter gene, vga(C), located on a multiresistance plasmid from a porcine methicillin-resistant Staphylococcus aureus ST398 strain. Antimicrob Agents Chemother 53:3589–3591. doi: 10.1128/AAC.00570-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kadlec K, Fessler AT, Couto N, Pomba CF, Schwarz S. 2012. Unusual small plasmids carrying the novel resistance genes dfrK or apmA isolated from methicillin-resistant or -susceptible staphylococci. J Antimicrob Chemother 67:2342–2345. doi: 10.1093/jac/dks235. [DOI] [PubMed] [Google Scholar]
- 29.Gómez-Sanz E, Kadlec K, Fessler AT, Zarazaga M, Torres C, Schwarz S. 2013. Novel erm(T)-carrying multiresistance plasmids from porcine and human isolates of methicillin-resistant Staphylococcus aureus ST398 that also harbor cadmium and copper resistance determinants. Antimicrob Agents Chemother 57:3275–3282. doi: 10.1128/AAC.00171-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bjorland J, Steinum T, Sunde M, Waage S, Heir E. 2003. Novel plasmid-borne gene qacJ mediates resistance to quaternary ammonium compounds in equine Staphylococcus aureus, Staphylococcus simulans, and Staphylococcus intermedius. Antimicrob Agents Chemother 47:3046–3052. doi: 10.1128/AAC.47.10.3046-3052.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.López M, Kadlec K, Schwarz S, Torres C. 2012. First detection of the staphylococcal trimethoprim resistance gene dfrK and the dfrK-carrying transposon Tn559 in enterococci. Microb Drug Resist 18:13–18. doi: 10.1089/mdr.2011.0073. [DOI] [PubMed] [Google Scholar]
- 32.Wendlandt S, Fessler AT, Kadlec K, van Duijkeren E, Schwarz S. 2014. Identification of the novel spectinomycin resistance gene spd in a different plasmid background among methicillin-resistant Staphylococcus aureus CC398 and methicillin-susceptible S. aureus ST433. J Antimicrob Chemother 69:2000–2003. doi: 10.1093/jac/dku067. [DOI] [PubMed] [Google Scholar]
- 33.Schijffelen MJ, Boel CH, van Strijp JA, Fluit AC. 2010. Whole-genome analysis of a livestock-associated methicillin-resistant Staphylococcus aureus ST398 isolate from a case of human endocarditis. BMC Genomics 11:376. doi: 10.1186/1471-2164-11-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paterson GK, Harrison EM, Murray GG, Welch JJ, Warland JH, Holden MT, Morgan FJ, Ba X, Koop G, Harris SR, Maskell DJ, Peacock SJ, Herrtage ME, Parkhill J, Holmes MA. 2015. Capturing the cloud of diversity reveals complexity and heterogeneity of MRSA carriage, infection and transmission. Nat Commun 6:6560. doi: 10.1038/ncomms7560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tong SY, Holden MT, Nickerson EK, Cooper BS, Koser CU, Cori A, Jombart T, Cauchemez S, Fraser C, Wuthiekanun V, Thaipadungpanit J, Hongsuwan M, Day NP, Limmathurotsakul D, Parkhill J, Peacock SJ. 2015. Genome sequencing defines phylogeny and spread of methicillin-resistant Staphylococcus aureus in a high-transmission setting. Genome Res 25:111–118. doi: 10.1101/gr.174730.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanczak-Mrozek KI, Manne A, Knight GM, Gould K, Witney AA, Lindsay JA. 2015. Within-host diversity of MRSA antimicrobial resistances. J Antimicrob Chemother 70:2191–2198. doi: 10.1093/jac/dkv119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.