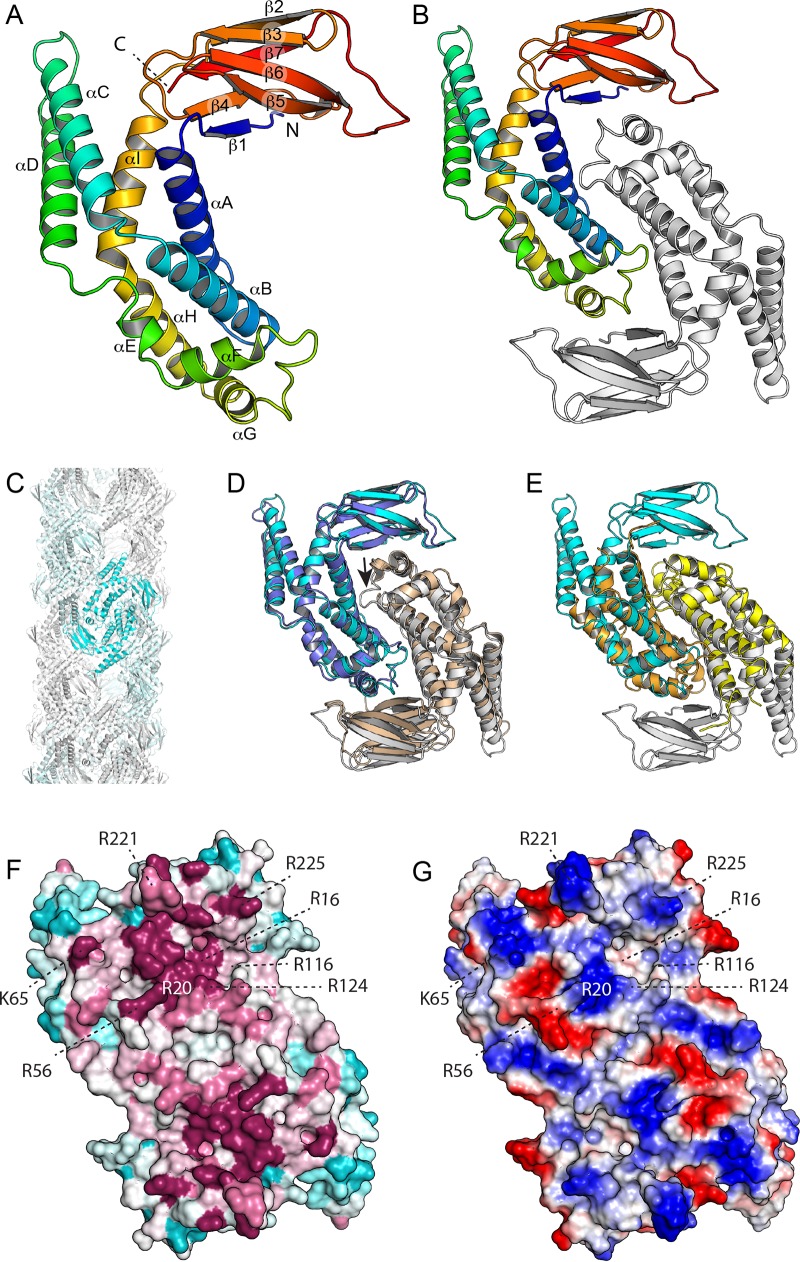
FIG 1.
Structure of E. coli ZapD. (A) The structure of the ZapD protomer is colored by secondary structure progression from the N (blue) to the C terminus (red). The ZapD structure is organized as a β-sandwich domain attached to an α-helical domain. Note that αC and αD protrude from the rest of the structure. (B) Structure of the ZapD dimer. (C) ZapD packing in the crystal results in large hollow fibers 110 Å in diameter. However, we find no evidence that this organization contributes to ZapD's function in vivo. (D) Superposition of E. coli ZapD (cyan and white) and ZapD from V. parahaemolyticus (blue and white; PDB entry 2OEZ). A single protomer (blue/cyan) was superimposed by DALI, with the second protomer positioned as it maps to the dimer interface. Note that in the V. parahaemolyticus ZapD structure, the αF-αG loop is largely disordered (small arrow). (E) Superposition of VpC_cass2 (PDB entry 3JRT; yellow/orange) dimer on the E. coli ZapD structure (cyan/white). Note that the VpC_cass2 structure superposes well on the core of the ZapD helical domain. (F) E. coli ZapD colored by sequence conservation score. A multiple-sequence alignment of ZapD homologs was mapped onto the ZapD structure using ConSurf. Highly conserved residues are shown in magenta, poorly conserved residues are in cyan, and residues with intermediate conservation are shown in white. (G) Electrostatic potentials are mapped onto the EcZapD surface. Blue indicates electropositive regions, and red indicates electronegative regions.