FIG 6.
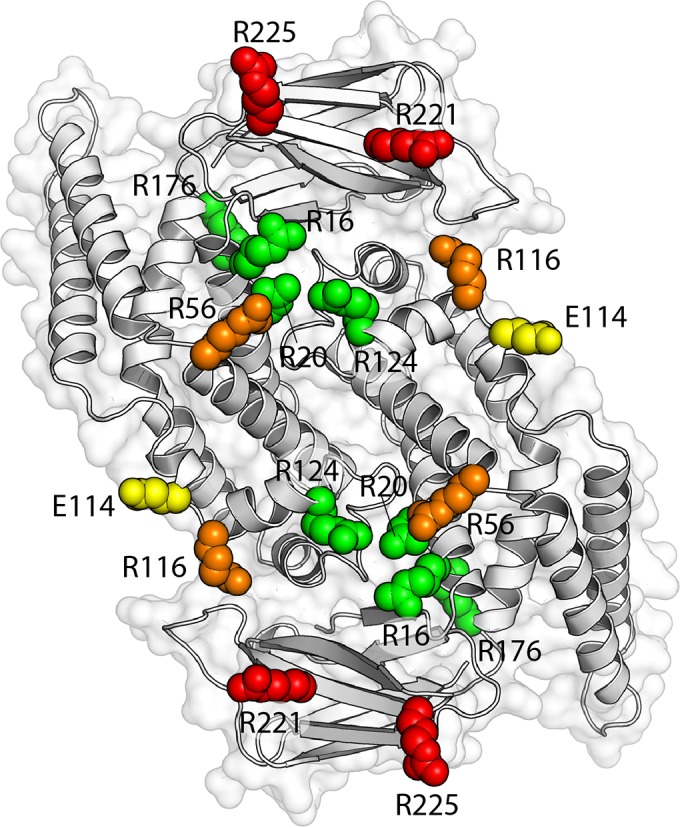
Mutant phenotypes mapped onto the EcZapD dimer structure. Orientation of the ZapD dimer as described for Fig. 1B, but here colored according to the effect of point mutations at each residue. Residues are grouped based on the results of sedimentation assays, bundle morphology, and cross-linking to FtsZ. Residues R16, R20, R124, and R176 are labeled in green. Mutation of these residues presented no significant deviations in EC50 or FtsZ bundle morphology compared to those of WT ZapD, and none of the residues form identifiable cross-links with FtsZ. Residue E114 is labeled in yellow. Mutation of E114 caused altered bundle morphology and a statistically significant increase in the number of FtsZ filaments per bundle compared to WT ZapD (16 versus 12). However, mutation of this residue did not affect the EC50, which was similar to that of WT ZapD (1.74 μM versus 1.86 μM), and E114 did not form identifiable cross-links with FtsZ. Residues R56 and R116 are labeled in orange. Mutation of R56 produced a statistically significant increase in EC50 (5.92 μM) and a statistically significant decrease in the number of FtsZ filaments per bundle compared to WT ZapD (7 versus 12), but R56 did not form identifiable cross-links with FtsZ. Mutation of R116 also demonstrated an increase in EC50 (2.77 μM) but demonstrated ineffective bundling of FtsZ. R116 also formed identifiable cross-links with FtsZ. Mutations in R221 and R225 are colored in red. Mutation of these residues produced a considerable increase in EC50s compared to that of WT ZapD (3.19 μM and 9.08 μM, respectively) and ineffective bundling of FtsZ filaments. R221 and R225 formed multiple identifiable cross-links with FtsZ.
