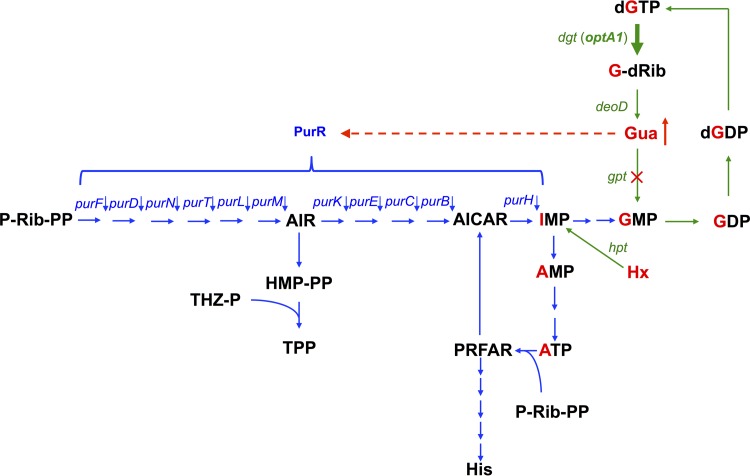
FIG 4.
Flow diagram of metabolic intermediates in purine biosynthesis. Green arrows indicate the enzymatic steps for turnover of the guanine moiety (labeled in red) within the guanine nucleotide pools, with the thick arrow indicating the elevated levels of dGTP triphosphohydrolase (dgt up-promoter allele in the optA1 strain). Also indicated in green is the Hpt-mediated salvage of hypoxanthine (Hx) to yield IMP. The blue arrows represent the mainstream de novo purine biosynthesis pathway, as well as its interconnections with histidine biosynthesis (from ATP and P-Rib-PP), resulting in AICAR production, and with thiamine biosynthesis (TPP) from the intermediate AIR. The red dashed line indicates the repressive effect of accumulated guanine (Gua) on the PurR-controlled genes (in the optA1 gpt strain). dgt, dGTP triphosphohydrolase gene; deoD, purine nucleoside phosphorylase gene; gpt, guanine phosphoribosyltransferase gene; hpt, hypoxanthine phosphoribosyltransferase gene; purR, purine repressor gene (encodes the transcription factor controlling de novo synthesis of purine nucleotides); Gua, guanine; Hx, hypoxanthine; G-dRib, deoxyguanosine; P-Rib-PP, 5′-phosphoribosyl-1-pyrophosphate (PRPP); AIR, 5′-aminoimidazole ribonucleotide; THZ-P, 4-methyl-5(β-hydroxyethyl)thiazol phosphate; HM-PP, 4-amino-5-hydroxymethyl-2-methylpyrimidine pyrophosphate; TPP, thiamine pyrophosphate; AICAR, 5-aminoimidazole-4-carboxamide ribonucleotide; PRFAR, phosphoribosyl formimino-5 aminoimidazol-4-carboxamide; His, histidine. The red cross indicates the lack of Gpt activity in the gpt deletion strain.