FIG 1.
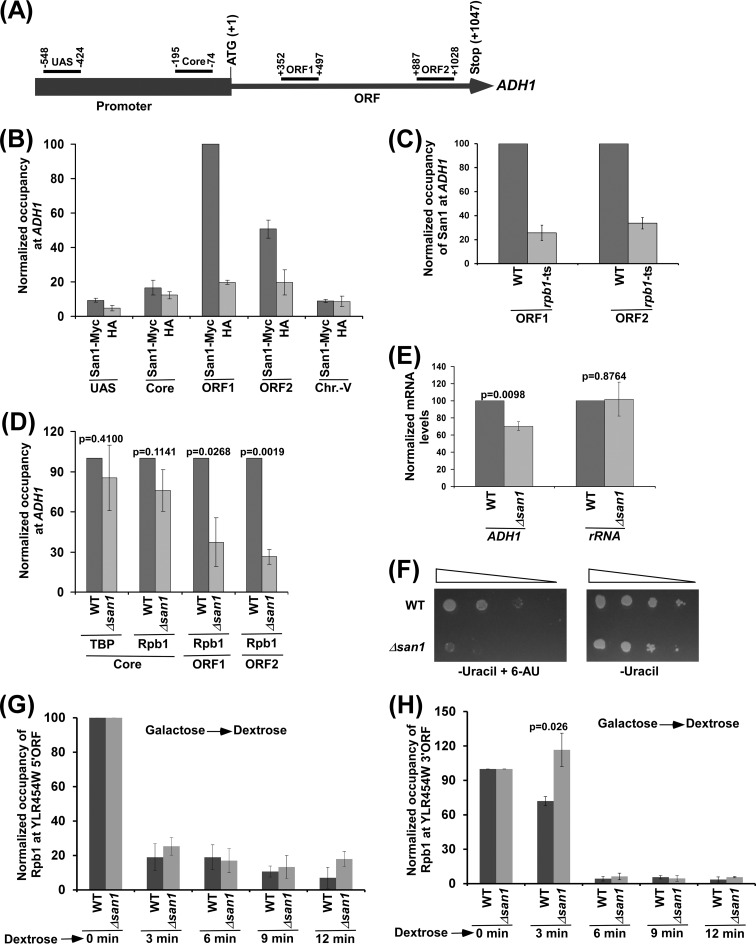
San1 associates with the coding sequence of ADH1 to promote the engagement of elongating RNA polymerase II into transcription. (A) Schematic diagram showing the locations of different primer pairs at ADH1 for the ChIP analysis. The numbers are presented with respect to the position of the first nucleotide of the initiation codon (+1). UAS, upstream activation sequence. (B) ChIP analysis of San1 association with ADH1. Yeast cells expressing Myc-tagged San1 were grown in YPD at 30°C to an OD600 of 1.0 prior to formaldehyde-based in vivo cross-linking. Immunoprecipitated DNA was analyzed by PCR using the primer pairs targeted to different locations of ADH1 and an inactive region within chromosome V (Chr.-V). The ratio of immunoprecipitate to input in the autoradiogram was measured. The maximum ratio was set to 100, and other ratios were normalized to 100. The normalized ratio (represented as normalized or relative occupancy or ChIP signal) is plotted in the form of a histogram. (C) San1 association with the ADH1 coding sequence is impaired in the rpb1-ts mutant following 1 h ts inactivation at 37°C. (D) ChIP analysis of the association of TBP and Rpb1 with ADH1 in the Δsan1 and wild-type strains, using an anti-TBP antibody against TBP and 8WG16 antibody (Covance) against the carboxy-terminal domain of Rpb1. (E) RT-PCR analysis of ADH1 mRNA levels in the Δsan1 and wild-type strains (with 18S rRNA as control). (F) Growth analysis of the Δsan1 and wild-type strains in the solid SC-uracil medium containing 2% dextrose with or without 6-AU (100 μg/ml) at 30°C. (G and H) ChIP analysis of Rpb1 association with 5′ (G) and 3′ (H) ends of the ∼8-kb-long YLR454W coding sequence (under the control of the GAL1 promoter) upon switching the carbon source in the growth media from galactose to dextrose. ORF, open reading frame.