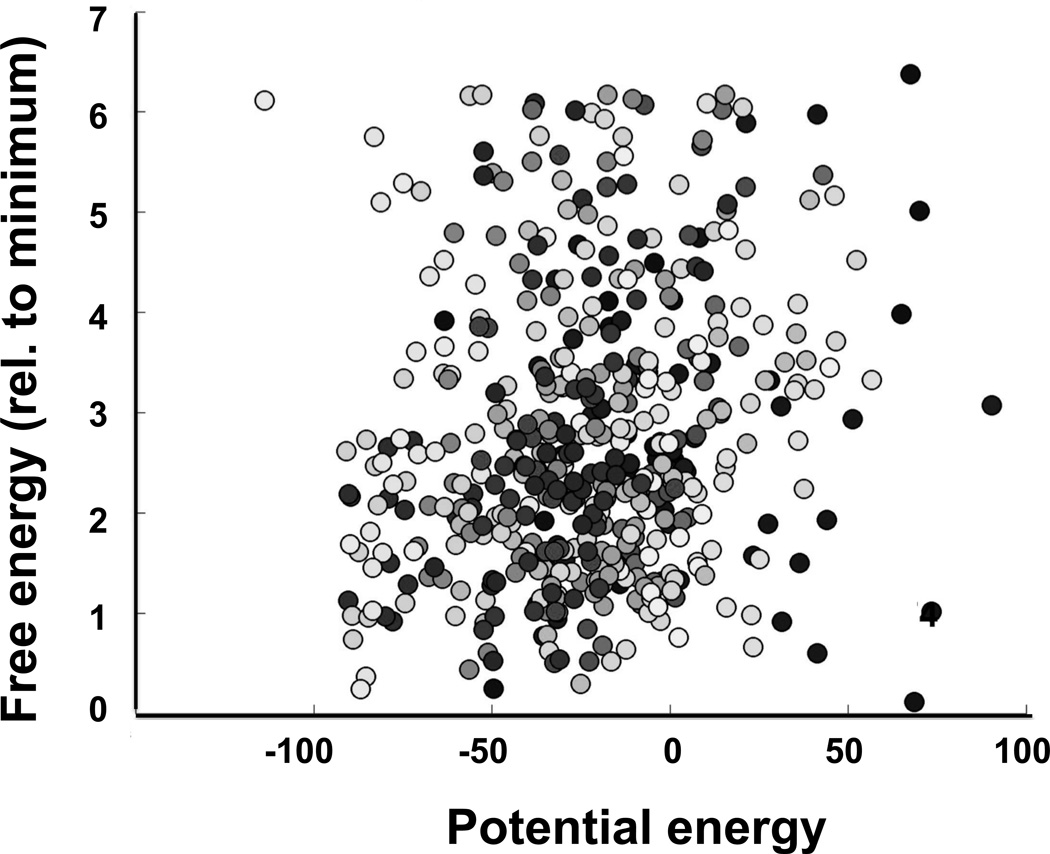
Figure 2.
Absence of a correlation between the relative free energy of the spin label and the potential energy of the labeled protein from Simulated Scaling Molecular Dynamics simulations. Free energy was calculated from frequency of visiting a specific xyz-voxel when the forcefields were fully switched on, potential energy was the lowest energy of any of those dynamic trajectory frames that corresponded to a given voxel. The correlation coefficient in this example was 0.14. Different shades of the symbols correspond to trajectories starting from different rotamers. Details of simulations including Charmm input files, spin label topology files can be found at http://biophysics.fsu.edu/fajer/.