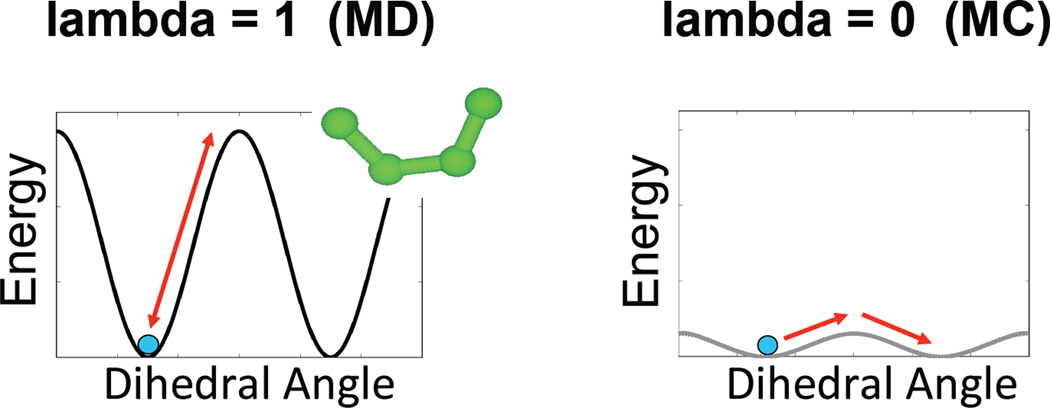
Figure 3.
The simulated scaling method applies a scaling factor (lambda) to the dihedral, electrostatics and van der Waals terms of the potential energy function. Here an example dihedral energy term is shown to illustrate the effect. At lambda of 1 the potential energy is unperturbed and regular molecular dynamics (MD) occurs. Sampling of different conformations can be frustrated by high energy barriers. At lambda of 0 the potential energy is completely flattened and transitions between conformations is limited only by diffusion, similar to the sampling observed in Monte Carlo (MC). However, this perturbation of the model is unphysical and results at lambda of 0 are not accurate. Only by moving back and forth through the entire 0 to 1 range of lambda can we achieve robust sampling of the accurate free energy surface.