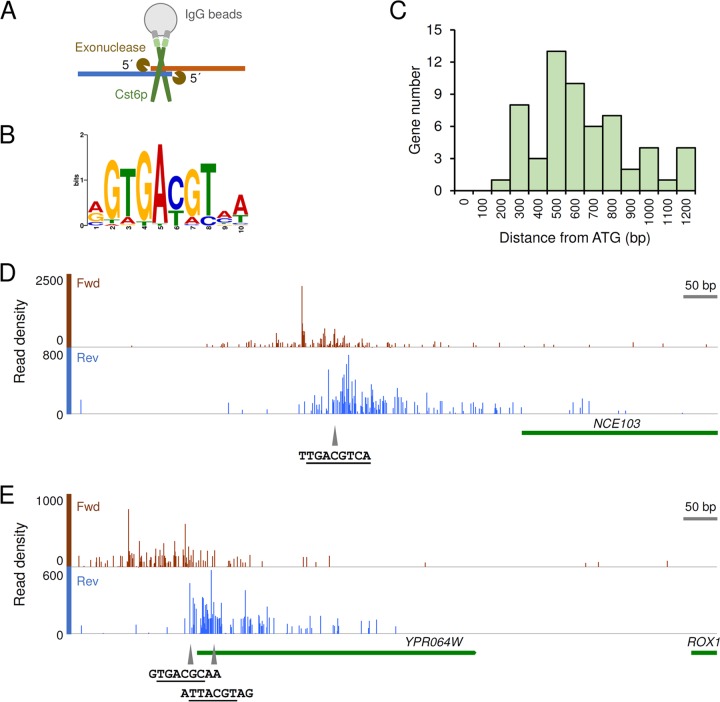
FIG 1 .
The identification of Cst6p binding sites by ChIP-exo. (A) Schematic representation of the ChIP-exo experiment. Cst6p with C-terminal tag is shown in the form of a homodimer. (B) Binding site motif enriched from the bound region sequences (E value = 8.9 × 10−10). (C) Distribution of the distances of binding events from the start codons of putative target genes. (D) Binding of Cst6p on the NCE103 promoter. (E) Binding of Cst6p on the ROX1 promoter. YPR064W is a dubious gene upstream from ROX1. Data shown in panels B to E are based on the binding events that occurred when cells were grown on ethanol. In panels D and E, the coverage graph showing the 5′ ends of reads was created from the aligned BAM file using “Depth Graph (Start)” in IGB (46). The putative binding sites of Cst6p are marked by triangles. The core binding sequences of the ATF/CREB family TFs (consensus sequence, 5′ TGACGT 3′) are underlined.