FIG 5 .
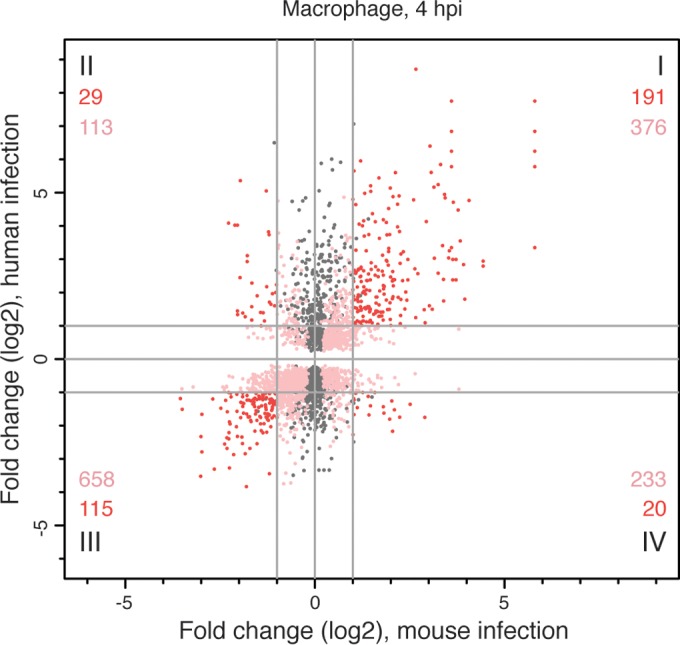
Responses of human and murine macrophages to L. major infection at 4 hpi after accounting for the phagocytosis effect. The differential expression profile of L. major-infected human macrophages was compared to that of L. major-infected murine macrophages collected at the same time point (40). Orthology mapping to mouse was done for the 3,273 human genes identified as differentially expressed at 4 hpi, after accounting for the phagocytosis effect, and the results were compared to the murine expression data set. A scatterplot showing the relationship between fold changes (log2 transformed) in mice (x axis) and humans (y axis) is shown, with each human-mouse gene pair represented by a single point. Some genes duplicates were introduced by the orthology mapping process. Points in gray represent genes in the human data set with an ortholog that was not significantly DE in the murine data set. Points in shades of red represent genes that were significantly DE in both data sets, with those showing <2-fold difference (log2 fold change, <1) in either/both host system(s) shown in pink and those with >2-fold difference (log2 fold change, >1) in both host systems shown in red. The numbers of unique genes represented by the red and pink points are indicated for each quadrant. The complete list of genes that were DE in both systems is provided in Data Set S4 in the supplemental material.
