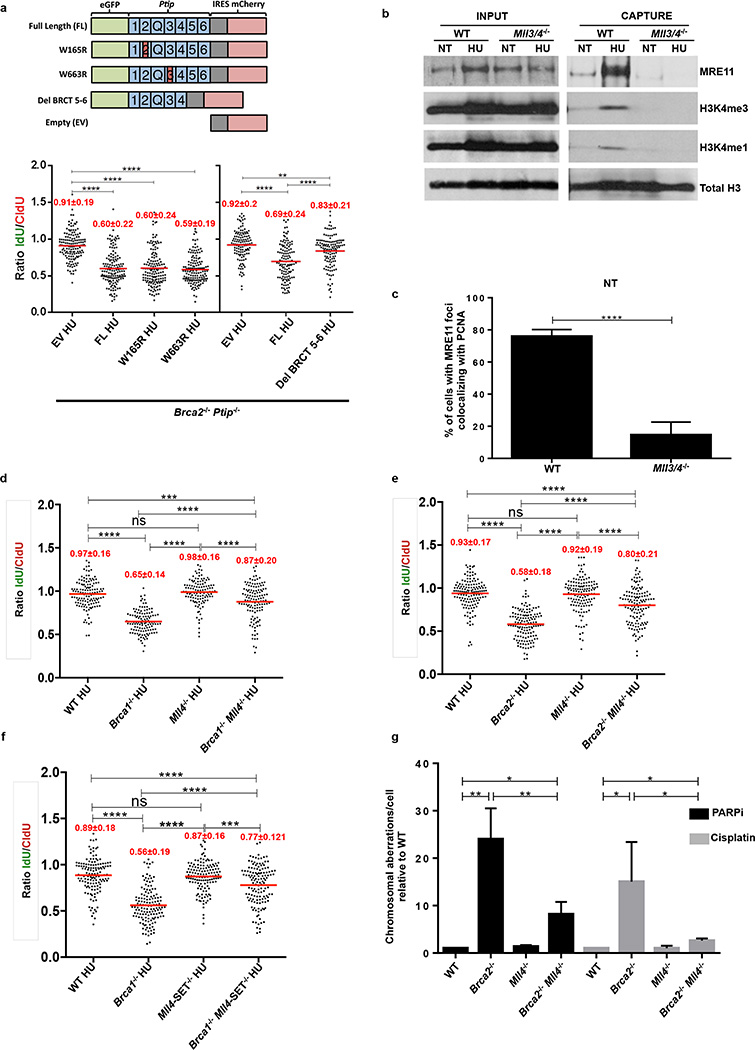
Extended Data Fig. 6. MLL3/4 promotes replication fork degradation in Brca2- deficient cells.
(a) Upper panel shows the schematic of the retroviral PTIP mutant constructs used to identify the domain of PTIP involved in driving replication fork degradation. Different BRCT domains in PTIP are numbered and Q represents the glutamine rich region between the 2nd and the 3rd BRCT domains. Lower graph panel shows the ratio of IdU vs. CldU upon HU treatment of Brca2−/− Ptip−/− B lymphocytes retrovirally infected with either EV, FL, W165R, W663R and Del BRCT 6-5 PTIP-mutant constructs and sorted for GFP or mCherry expression. Numbers in red indicate the mean and standard deviation for each sample (**P ≤ 0.01, ****P ≤ 0.0001, Mann-Whitney test). 125 replication forks were analyzed for each condition. (b) WT and Mll3/4−/− MEFs were EdU-labeled for 15 min and treated with 4 mM HU for 4 hr. Proteins associated with replication forks were isolated by iPOND and detected by Western blotting with the indicated antibodies. (c) Quantification of the percentage of cells with MRE11 foci co-localizing with PCNA in late S-phase in WT and MLL3/4−/− MEFs (n=150 cells analyzed). Experiments were repeated 3 times. (d-f) Ratio of IdU vs. CldU upon HU treatment in WT, Brca1−/−, Brca2−/− Mll4−/−, Mll4-SET−/−, Brca1−/−Mll4−/−, Brca2−/−Mll4−/− and Brca1−/−Mll4-SET−/− B cells. Numbers in red indicate the mean and standard deviation for each sample (ns, not significant, ****P ≤ 0.0001, Mann-Whitney test). At least 125 replication forks were analyzed for each genotype. (g) Genomic instability measured in metaphase spreads from splenic B cells derived from WT, Brca2−/−, Mll4−/−, Brca2−/−Mll4−/− B cells treated overnight with 1 µM PARPi or with 0.5 µM cisplatin (**P ≤ 0.01, ***P ≤ 0.001, Unpaired t test). 50 metaphases were analyzed per condition. Experiments were repeated 3 times.