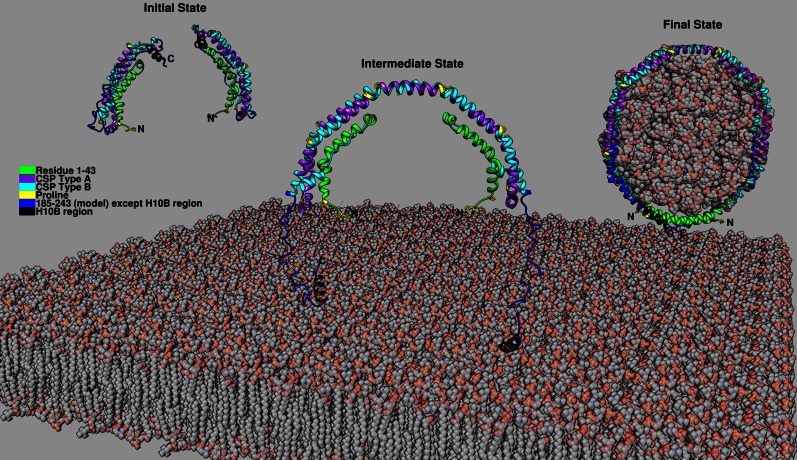
Fig. 7.
Proposed full-length apoA-I in the lipid-free state and a possible three-state mechanism of HDL formation. Full-length apoA-I in lipid-free state: residues 1-184 from Δ(185-243) crystal structure (Protein Data Bank accession number: 3R2P), residues 185-243 from Δ(1-43) crystal structure (Protein Data Bank accession number: 1AV1) with relaxation from helix to random coil except the H10B region. Initial state: Monomeric apoA-I adopts the helix bundle conformation with the C-terminal helical H10B region forming exposed hydrophobic surface. Intermediate state: Two monomeric apoA-I close enough to facilitate the domain swap interaction and result in dimerization due to binding to lipids spontaneously or facilitated through ABCA1. Final state: Lipids are solubilized by the double belt dimer with additional helical conformation forming in the unstructured region of the C-terminal domain further stabilizing the interaction and resulting in the release of discoidal HDL from the cell membrane.