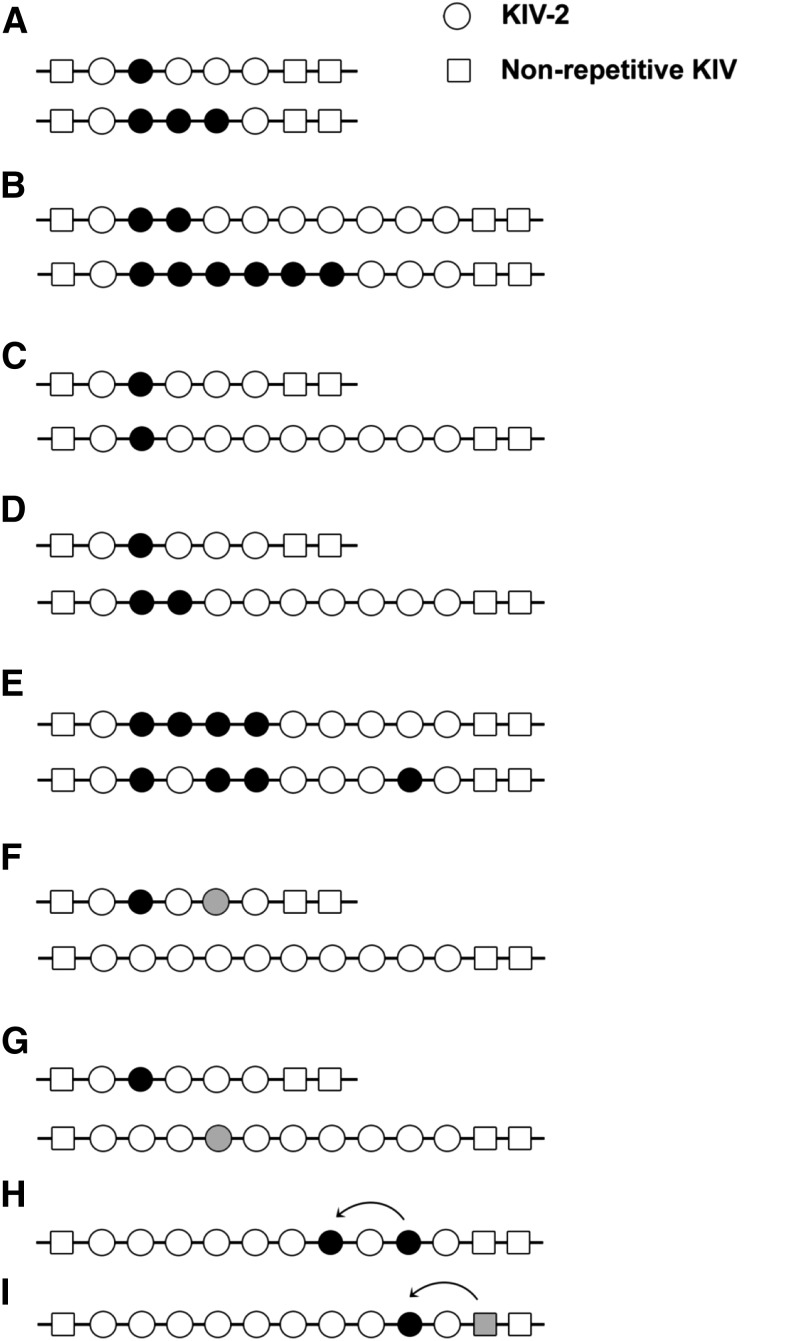
Fig. 5.
Possible distributions of sequence variations in the KIV-2 CNV. Different scenarios for the distributions of sequence variants (shown as filled circles) in KIV-2 copies of different sizes are illustrated. Low and high intra-allelic frequencies of a variant on short alleles (A) and longer alleles (B). C: The same number of KIV-2 copies harbors the variant on a short and a longer allele, i.e., the intra-allelic frequency of the variant is higher on the short allele. Thus detection of the variant is more likely if present on the short allele. D: Both alleles have the same intra-allelic frequency (20%), though the number of copies carrying the variant is different. Hence the probability of detection is the same. The order of variants carrying KIV-2 copies within the allele might vary, as shown in (E). These scenarios cannot be distinguished by present methods. Different variants can be allocated in cis (F) (shown for a genotype with one short and one longer allele) or in trans (G). While scenarios (F) and (G) cannot be distinguished in analyses based on diploid samples, this is possible if analysis is based on separated alleles. Panels H and I depict possible spreads of variants across the KIV-2 CNV (H) and between KIV-2 copies and the neighboring nonrepetitive KIV-3 (I) by gene conversion. Figure used and modified with permission from (200).