Figure 1.
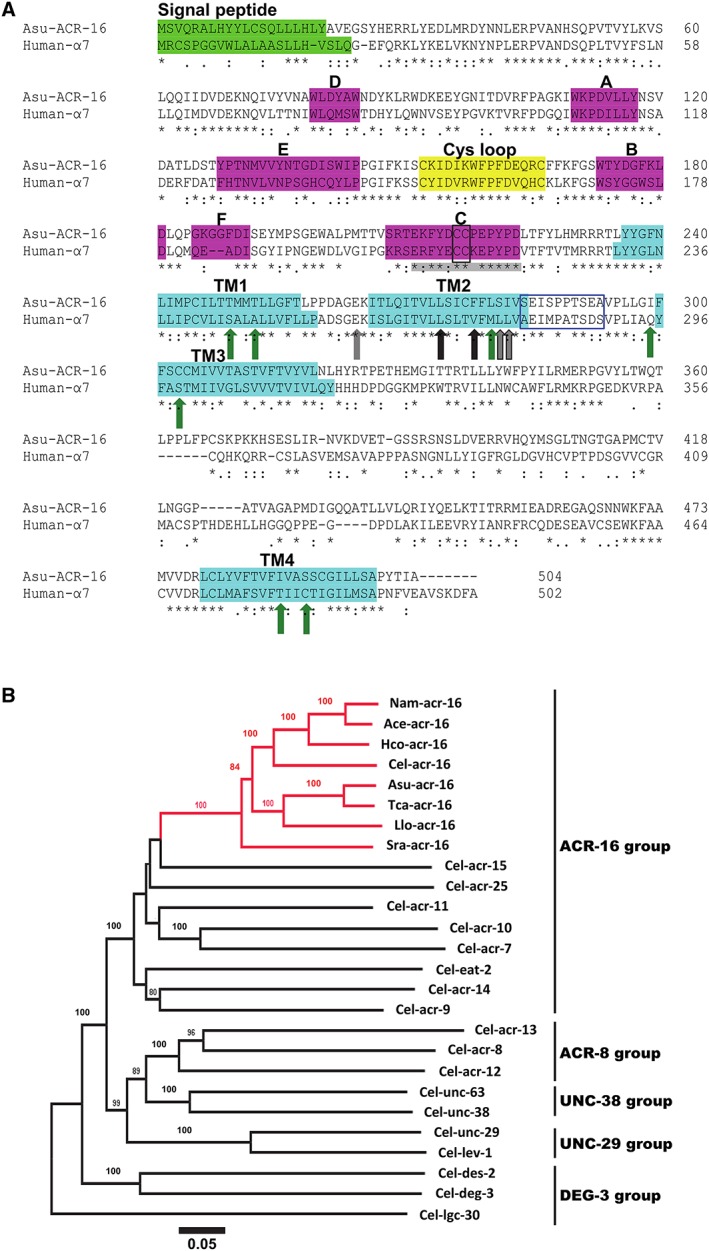
(A) Amino acid sequence alignment of Asu‐ACR‐16 and human‐α7 nAChR subunits. The signal peptide (bright green box), ACh‐binding loops A–F (pink boxes), cys‐loop (yellow box) and transmembrane regions TM1–TM4 (turquoise boxes) are indicated. The vicinal cysteines (black‐edged box) that characterize an α‐subunit are present in the C‐binding loop. The blue‐edged box between TM2 and TM3 represents the region where PNU120596 acts on α7 nAChRs. Green arrows are residues important for positive allosteric modulation of α7 receptors by ivermectin. Grey (and grey outline) arrows are residues important for permeability of α7 receptors to Ca2 +. Black (and black outline) arrows are residues affecting α7 receptor desensitization. Residues in C‐binding loop of α7 nAChRs that bind α‐BTX are highlighted in grey. (B) Distance tree showing relationships of ACR‐16 homologues in parasitic nematode species with AChR subunit sequences from C.elegans. A neighbour joining tree was generated with deduced amino acid sequence from AChR subunits representative from the ACR‐16, ACR‐8, UNC‐38, UNC‐29 and DEG‐3 group as defined by Mongan et al., (1998). Three letter prefixes in AChR subunit names: Ace, Asu, Cel, Hco, Llo, Nam, Sra and Tca, refer to A. ceylanicum, A. suum, C. elegans, H. contortus, L. loa, N. americanus, S. ratti and T. canis respectively. ACR‐16 orthologues are highlighted in red. Numbers at each branch indicate percentage bootstrap values (>80%) corresponding to 1000 replicates. The scale bar represents substitutions per site. The Cel‐lgc‐30 subunit sequence was used as an outgroup.
