Fig. 3.
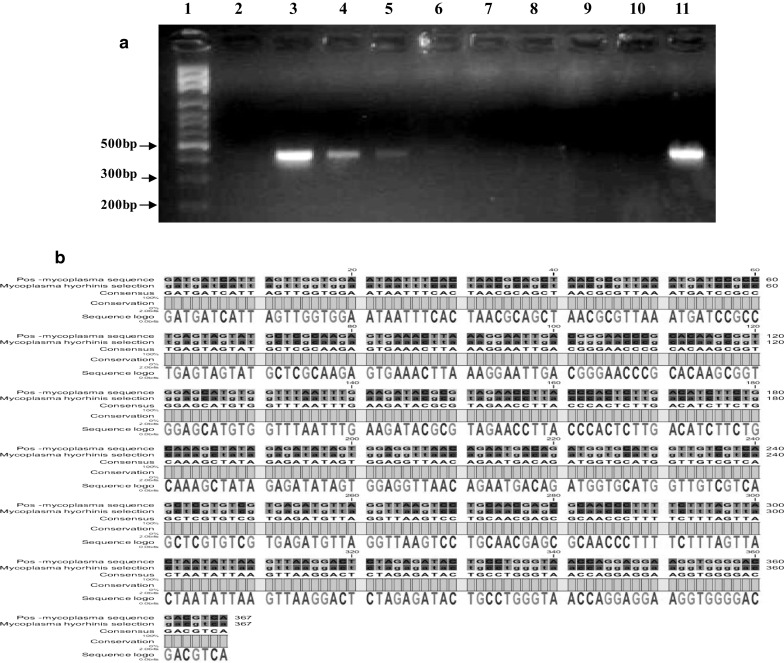
a Determination of the limit of detection for the PCR assay. Gel electrophoresis results of optimized PCR assay performed with universal primers on different serial dilutions of DNA obtained from a culture of M. hyorhinis. Lane 1 DNA Size marker (100 bp DNA Ladder, Roche XIV), lane 2 DNA-free water (negative control), lane 3 3 ng (positive), lane 4 300 pg (positive), lane 5 30 pg (positive), lane 6 3 pg (negative), lane 7 300 fg (negative), lane 8 30 fg (negative), lane 9 3 fg (negative), lane 10 300 atg (negative), lane 11 Undiluted-DNA strain of M. hyorhinis as a positive control (30 ng). b The alignment of the sequence of PCR product obtained by the amplification of M. hyorhinis 16S rRNA with the corresponding reference sequence at the NCBI Gene Bank data base. This comparison confirmed that the universal primers used in the study specifically amplified the desired fragment of a conserved sequence in all mycoplsma species
