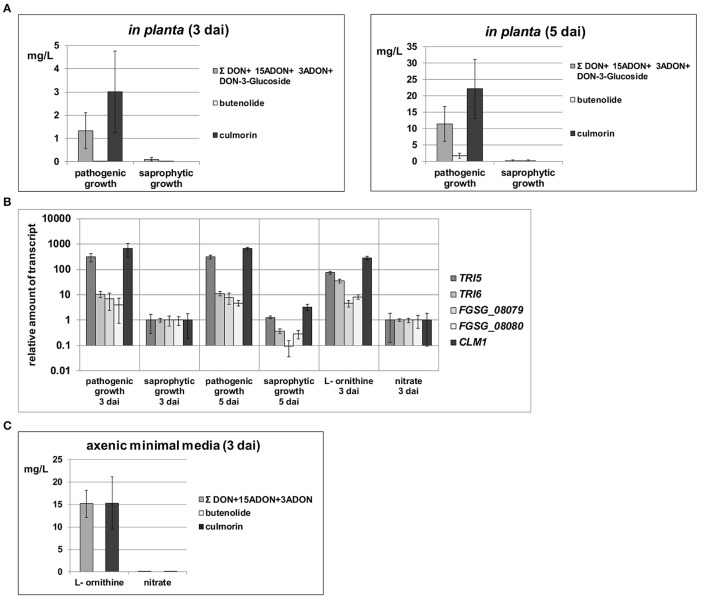
Figure 3.
Quantification of total DON/DON derivatives as well as butenolide and culmorin levels in different sample types and RT-qPCR analysis of key enzyme expression levels in respective SM gene clusters. (A) In planta cell extracts were analyzed for DON and its derivatives (15ADON, 3ADON, and DON-3-Glucoside) as well as for butenolide and culmorin 3 and 5 days after inoculation (dai). (B) Relative transcription levels normalized to β-tubulin transcription are shown for TRI5 (trichodiene synthase) and TRI6 (zinc finger transcription factor) of the core trichothecene biosynthesis cluster, FGSG_08079 (predicted cytochrome P450 benzoate 4- monooxygenase) and FGSG_08080 (putative regulatory protein containing a zinc finger motif) of the butenolide synthesis cluster and CLM1 encoding a longiborneol synthase necessary for culmorin biosynthesis. Levels were analyzed in planta during pathogenic (living plant) and saprophytic (dead plant) growth as well as in axenic cultures. Within the in planta samples expression levels of all tested genes on saprophytic material were arbitrarily set to 1 to allow direct comparison between these samples. For axenic samples, expression levels found on nitrate were arbitrarily set to 1 to allow comparisons of these in vitro samples. For all genes and conditions, comparative fold transcription levels are depicted on a logarithmic scale. (C) Filtrates of axenic cultivations on L- ornithine and nitrate harvested 3 dai were analyzed for DON and its derivatives (15ADON, 3ADON) as well as for butenolide and culmorin.