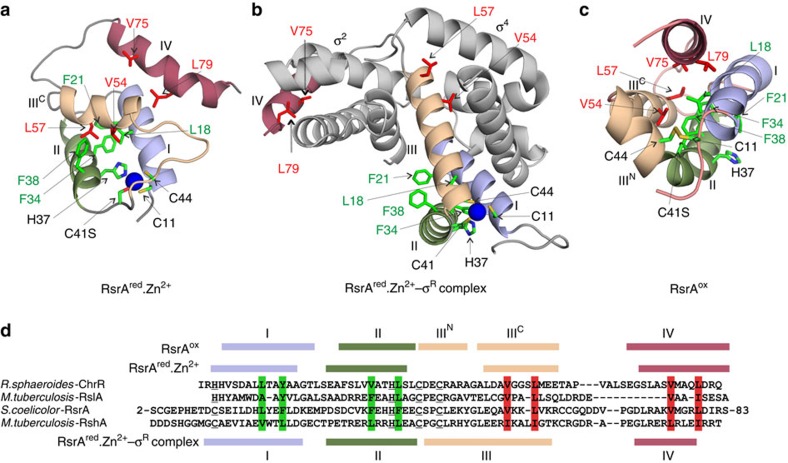
Figure 7. RsrAred.Zn2+ uses hydrophobic core residues to bind σR that are sequestered to the RsrAox interior following oxidation.
(a) Solution structure of RsrAred.Zn2+. RsrA helices are coloured N to C as in the sequence alignment in d. Zinc is shown as a blue sphere and zinc ligands coloured by atom type. Conserved or conservatively substituted hydrophobic residues that contribute to RsrA's hydrophobic core in all three of its structural states (a–c) are coloured green, while those that also interact with σR are coloured red. (b) Homology model of the RsrAred.Zn2+–σR complex validated by homo-bifunctional lysine-specific crosslinking (Supplementary Fig. 5). The structure of RsrAred.Zn2+ changes markedly to embrace σR. (c) Solution structure of RsrAox where the trigger disulfide is formed between residues Cys11 and Cys44, expelling bound zinc and repacking the hydrophobic core. (d) Sequence alignment of RsrA and other ZAS proteins (ChrR, RshA and RslA). Zinc ligands in each protein are underlined. Helices in all three structural forms of RsrA are coloured as in a–c. Vertical green shading shows conserved hydrophobic residues that contribute to the hydrophobic core of RsrA in all three structural states (RsrAred.Zn2+, RsrAred.Zn2+–σR complex and RsrAox). Vertical red shading shows conserved hydrophobic residues in RsrA that contribute to the hydrophobic cores of RsrAred.Zn2+ and RsrAox, but also contribute to the protein–protein interface in the RsrAred.Zn2+–σR complex.