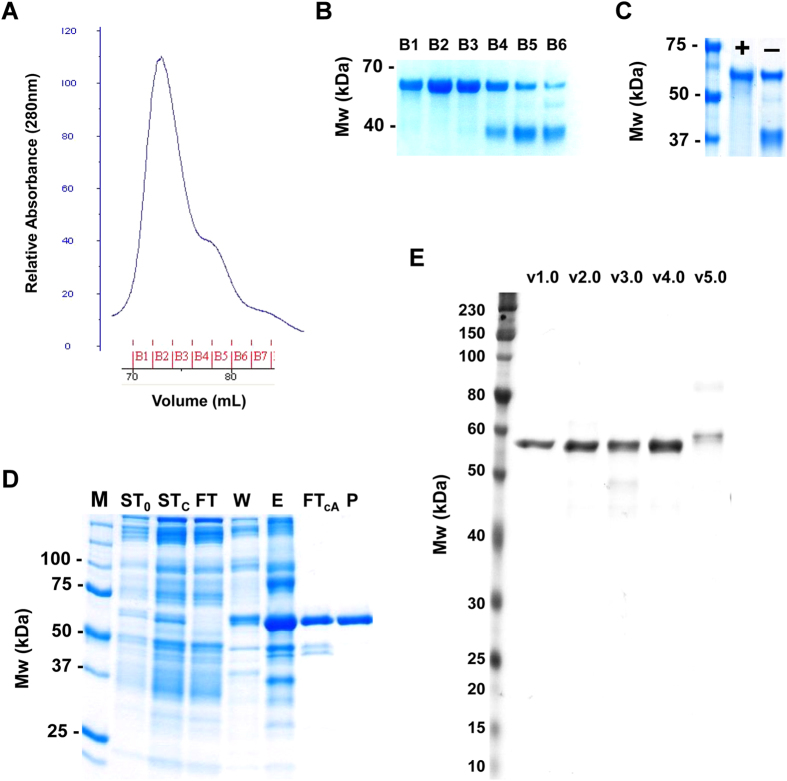
Figure 2. Purification of PfRH5 protein variants.
(A) Size exclusion chromatography (SEC) curve of the Ni-IMAC peak fraction pool run on Superdex200 in 20 mM HEPES pH 7.4, 150 mM NaCl. The PfRH5 peak and the shoulder of contaminant 38 kDa Drosophila S2 protein are shown. (B) Coomassie gel analysis of SEC fractions. 10 μL of the SEC fractions B1–B6 were resolved on a 12% Tris-Glycine gel and proteins were stained with InstantBlue. (C) Coomassie gel analysis of pools of PfRH5 SEC peak fractions showing product purified in the presence (+) or absence (−) of the Con A 4B Sepharose purification step. Proteins were resolved on a 12% Tris-HEPES gel and stained with InstantBlue. (D) Coomassie gel analysis of a representative purification run of PfRH5 v1.0. Proteins were separated on a Bio-Rad Any kD TGX gel and stained with SimplyBlue SafeStain. M: molecular weight markers; ST0: starting material; STC: starting material after TFF and buffer exchange; FT: flow-through from HisTrap column; W: HisTrap 40 mM Imidazole wash; E: Elution from HisTrap with 400 mM Imidazole; FTcA: flow-through from Con A column; P: final product after SEC. (E) Coomassie gel analysis of all purified PfRH5 protein variants run head-to-head (v1.0–v5.0). 1.25 μg each protein sample (DTT reduced and heat denatured) was separated on a Bio-Rad Any kD Criterion TGX gel and stained with InstantBlue.