Fig. 1.
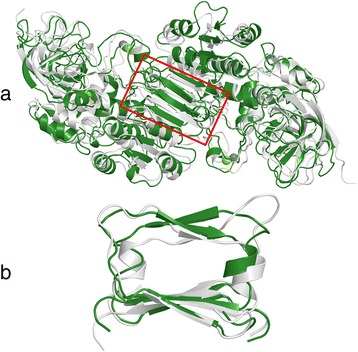
Comparison of dimers of class Iγ ADH and class V ADH after 20 ns of molecular dynamics simulations. The enzyme models of class Iγ ADH (white) and class V ADH (green) have a high level of structural similarity. a: The full dimers of class Iγ ADH and class V ADH after 20 ns of molecular dynamics simulations. b: The dimer interaction region (position 282–320) marked in red in A, rotated 90°. It contains a short β-α motif. The α-helix was not identified by DSSP after two out of three molecular dynamics simulations of the class V ADH model; one such run, with non-α-helical Ramachandran angles, is shown here. This tendency was not observed in any other ADH models (e.g. class Iγ ADH, white), and only in mouse class II ADH among the structures from multicellar organisms available in the RCSB PDB
