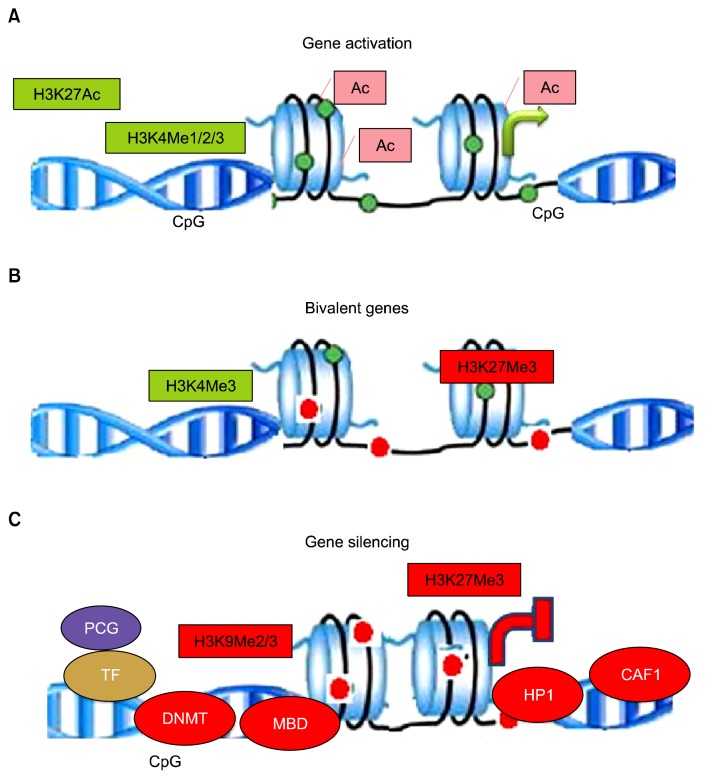
Fig. 2.
Histone modifications: Active genes: Open chromatin structure of transcriptionally active gene with loosely spaced nucleosomes. Acetylation of lysine neutralizes the positive charge, reducing affinity between histone and DNA, which functions as platforms for the recruitment of transcription factors or chromatin remodelers, thus histone modifications directly effects nucleosomal architecture. H3K4me3 is enriched around transcription start sites. H3K4me1 is enriched around enhancers and downstream. H3K27ac is enriched around active enhancers and transcription start sites. Bivalent genes: In undifferentiated stem cells, both H3K4me3 and H3K27me3 (active and inactive marks, respectively) are enriched around transcription start sites on many genes. The multiple coexisting histone modifications are associated with activation and repression. Inactive genes: H3K9me3 is broadly distributed on inactive regions. H3K27me3 and H3K9me3 are usually not colocalized. Proteins associated for transcription silencing are DNMT- DNA methyl-transferase, MBD-Methyl-binding domain, HP-1-Heterochromatin protein and CAF-1-Chromatin assembly factor1.