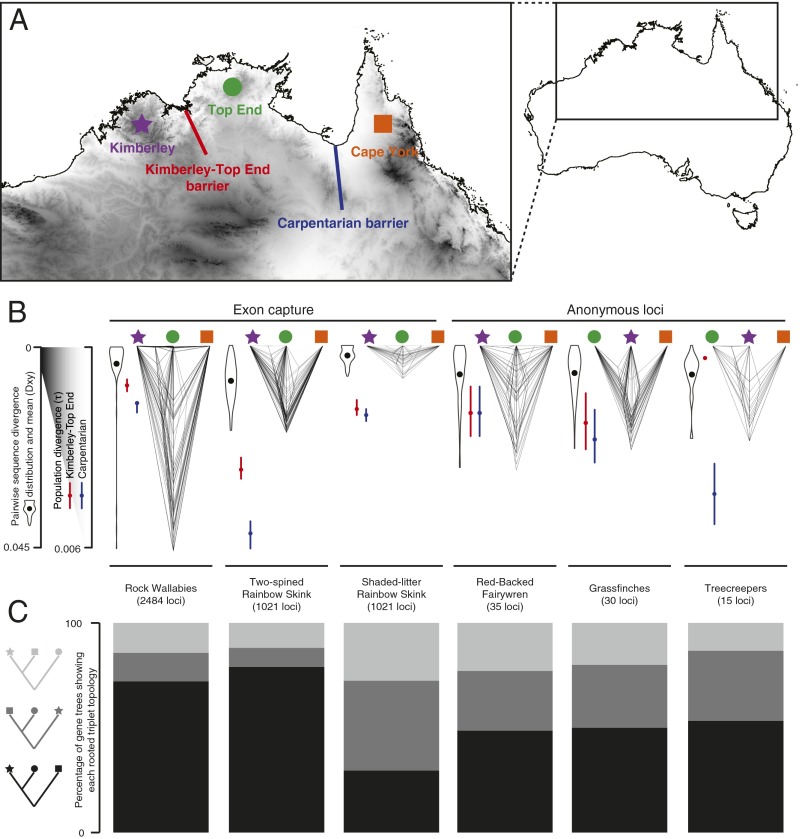
Fig. 3.
Gene tree heterogeneity in multilocus phylogeographic datasets of birds (Red-Backed Fairywren, Malurus melanocephalus; Poephila grassfinches; Climacteris treecreepers), skinks (Two-Spined Rainbow Skink, C. amax; Shaded-Litter Rainbow Skink, C. munda), and mammals (Petrogale rock-wallabies) across northern Australia. (A) Map of northern Australia showing the KTEB and CB that separate the KIM, TE, and CY faunas. (B) Cloudograms illustrate topological and branch length variation of gene trees. Violin plots represent the distribution of pairwise sequence divergences across the CB, and black dots indicate mean pairwise sequence divergence, or Dxy. Red dots and lines are estimates and 95% confidence intervals of population divergence across the KTEB, whereas green dots and lines are estimates and 95% confidence intervals of population divergence across the CB. (C) Distribution of rooted triplets shows that gene trees exhibiting deeper divergence times across the CB than the KTEB are the most frequent in all taxa except the Shaded-Litter Rainbow Skink. Additional details are provided in SI Text.