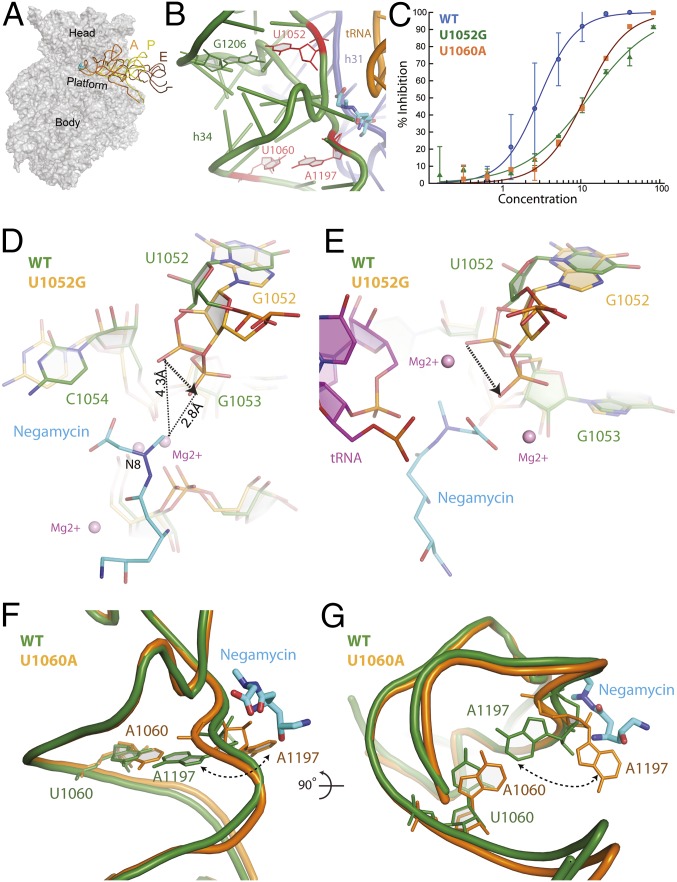
Fig. 1.
Negamycin resistance mutations. (A) Negamycin binds near the A site on the head domain of the 30S subunit. A, P, and E represent bound tRNA molecules. (B) Negamycin resistance mutations U1052, U1060 (11, 12), and A1197 (12) are positioned on h34 and in close proximity to the negamycin binding site. Previously identified resistance mutations are colored in red. (C) Translation activity of wild-type (blue circles; IC50 value of 2.9 µM), U1052G (green triangles; IC50 value of 13 µM), and U1060A (orange squares; IC50 value of 11 µM) ribosomes in the presence of negamycin in a luminescence-based in vitro-coupled transcription–translation assay with purified components. (D) Mutation U1052G significantly distorts the phosphate backbone of h34. The U1052G mutant (orange) has been superimposed on the wild-type E. coli structure (green) bound to negamycin. (E) Superposition of the U1052G mutant from E. coli (orange) on the mRNA–tRNA–negamycin complex from T. thermophilus (green) (12). (F) Acquisition of the U1060A mutation causes a localized perturbation that drives nucleotide A1197 into a flipped out conformation. The U1060A mutant structure (orange) has been superimposed on the wild-type E. coli structure bound to negamycin (green). (G) This panel is rotated 90° along the horizontal axis, illustrating the steric clash with negamycin.