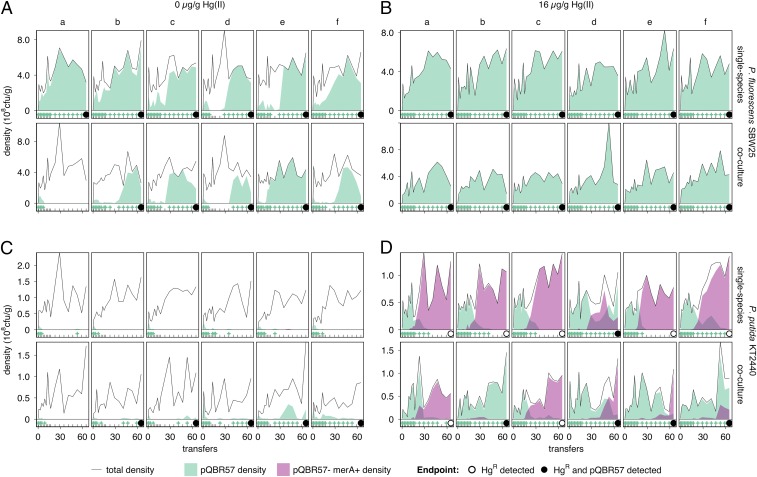
Fig. 1.
Coculture with favorable host P. fluorescens promotes plasmid carriage in unfavorable P. putida. (A) P. fluorescens populations evolved with 0 µg/g Hg(II). The upper row of subpanels shows single-species populations; the lower row shows populations cultured alongside P. putida (coculture). Six replicate populations (columns, labeled a–f) were initiated for each treatment. Each subpanel shows, for an individual population, total density at transfer (solid line), the density of pQBR57+ (filled green area below the line), and the density of pQBR57– merA+ mutants (filled purple area below the line). For clarity, tick marks at the bottom of each subpanel indicate sampling times, and green “+” symbols indicate detection of pQBR57. A black circle at the final sampling point (transfer 65) indicates that HgR remained in the population at the end of the experiment; filled circles indicate pQBR57 (and HgR) remained. Note that no pQBR57– merA+ mutants were detected in P. fluorescens. (B) P. fluorescens populations evolved with 16 µg/g Hg(II). As in A, except evolved with 16 µg/g Hg(II). (C) P. putida populations evolved with 0 µg/g Hg(II). As in A, except populations were P. putida. The lower row of subpanels shows populations cultured alongside P. fluorescens (coculture). Each population of cocultured P. putida a–f was grown with the corresponding cocultured P. fluorescens population (a–f, A). (D) P. putida populations evolved with 16 µg/g Hg(II). As in C, except evolved with 16 µg/g Hg(II). Different y-axis scales are used for each species: P. fluorescens density was ∼5x P. putida.