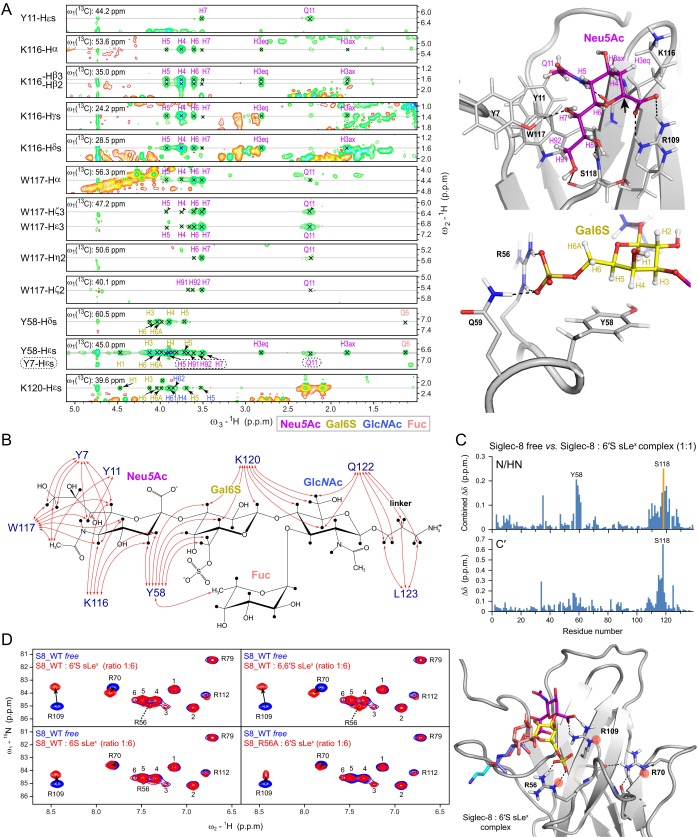
Fig. S4.
Intermolecular NOEs between Siglec-8 and 6′S sLex and arginine residues involved in Siglec-8-glycan recognition, detected by solution NMR spectroscopy. (A) (Left) The 3D 13C F1-edited F3-filtered NOESY spectrum of 13C/15N-labeled Siglec-8 in complex with unlabeled 6′S sLex, recorded at 1H frequency of 700 MHz, at 303 K. Depicted are exemplary sections of indicated Siglec-8 resonances at ω2 (13C edited/selected) showing clear intermolecular NOEs to indicated carbohydrate resonances, which appeared well-dispersed in the direct dimension ω3 (13C filtered/suppressed). (Right) Close-up views of the recognition of Neu5Ac (Top) and the Gal6S (Bottom) in the Siglec-8–6′S sLex complex structure, displayed in cartoon representation. Side chains of key protein residues and the carbohydrate residues are represented as sticks and labeled. Positions of the NMR-observable carbohydrate 1H atoms are indicated with labels and colored by residue type. The black arrow points to the anomeric carbon (C2) atom of Neu5Ac to which the underlying carbohydrate moieties (omitted for visual clarity) are linked. (B) Graphical summary of intermolecular NOEs observed between 13C/15N-labeled Siglec-8 and unlabeled 6′S sLex, detected in 2D 13C/15N F2-filtered NOESY and 3D 13C F1-edited F3-filtered NOESY spectra as described in Materials and Methods, schematically illustrated by red arrows onto the chemical structure of 6′S sLex. (C) Chemical shift differences between free Siglec-8 and Siglec-8 in complex with 6′S sLex (at molar ratio of 1:1), plotted as a function of residue number. (Top) Combined 1H/15N backbone amide chemical shift differences derived from 2D 1H,15N-HSQC spectra. (Bottom) The 13C backbone carbonyl chemical shift differences derived from 3D HNCO spectra. The orange bar indicates that the Ser118 amide resonance peak was invisible in the complex as a result of severe line broadening, due to intermediate exchange effects. (D) (Left) The 2D 1H,15N-HSQC spectra of 15N-labeled Siglec-8 wild type (S8_WT) in its free form (blue contours), overlaid with spectra of indicated Siglec-8/glycan ligand complexes (red contours), showing the spectral regions of the arginine side chain Nε-Hε resonances. The used Siglec-8 lectin domain expression construct contains in total 11 arginine residues (including the nonnatural C-terminal Arg145). In the 2D 1H,15N-HSQC spectrum of free Siglec-8 wild type only 10 Nε–Hε cross-peaks are visible/resolved. Nε-Hε cross-peaks that could be sequence-specifically assigned are labeled accordingly, whereas unassigned resonances, corresponding to those of Arg5, Arg96, Arg99, Arg101, Arg145, are numbered arbitrarily by 1–6. The Nε-Hε cross-peaks of the conserved essential Arg109 and the one of Arg70 show chemical shift perturbations in all four complexes. In contrast, the Arg56-Nε-Hε cross-peak, which is invisible (or overlapped) in the 1H,15N-HSQC spectrum of free Siglec-8, appears only in complexes of Siglec-8 wild type with 6′-sulfated-sLex variants (sulfo-group at the C6 position of Gal) but remains invisible/unaffected in complexes with 6-sulfated (at the C6 position of GlcNAc) sLex variants, as well as in the complex of the Siglec-8 R56A mutant with 6′S sLex. These observations strongly indicate that the guanidinium group of Arg56 is directly involved in the recognition of the sulfate group at the Gal C6, present only in the 6′S sLex and 6,6′S sLex tetrasaccharides. All spectra were recorded at 10 °C, in NMR buffer at pH 7.4 (5% D2O). The observed pattern of arginine Nε-Hε chemical shift changes can be explained by the NMR solution structure of the Siglec-8:6′S sLex complex, as shown in Right, which shows a cartoon representation of the Siglec-8: 6′S sLex complex, showing the side chains of Arg56, Arg70, and Arg109 as sticks, and their Nε-Hε amide atoms are marked by red circles: the essential Arg109 forms a salt bridge (end-on) with the carboxyl group of the terminal Neu5Ac. Arg70 (C′D loop) makes no direct contacts to the carbohydrate ligand but forms intraprotein hydrogen bonds to the backbone carbonyl oxygens of Pro57 and Asp60 on the CC′ loop (shown in Fig. S3 C and D). Arg56 (CC′ loop) forms a salt bridge (side-on) with the sulfate group at the C6 position of the Gal moiety. NH2 resonances were degenerated or not visible and could not be assigned except for Arg79 (see Fig. S3E.)