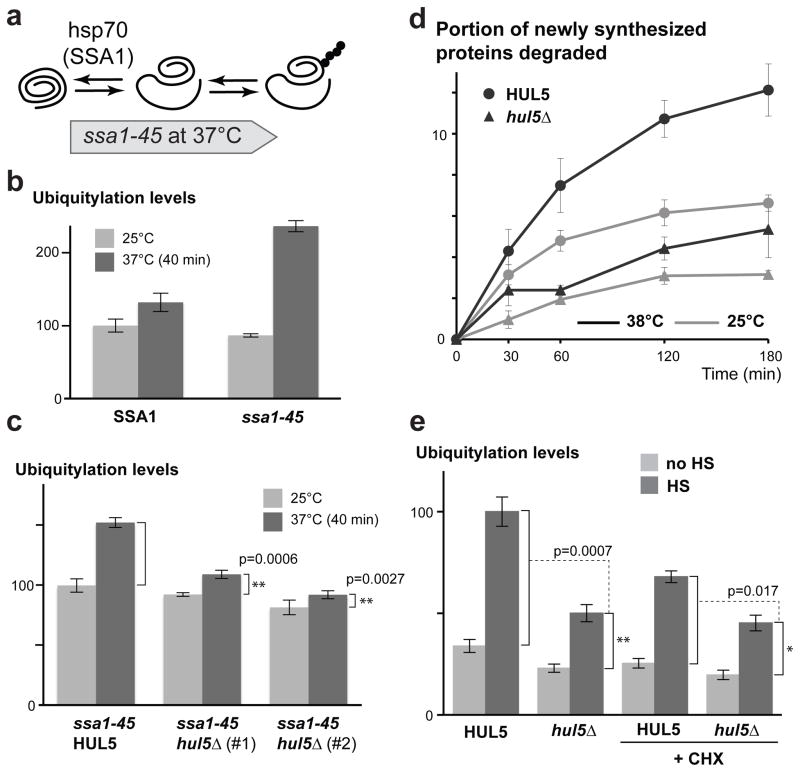
Figure 5.
HUL5 is essential for the ubiquitylation of proteins misfolded in absence of SSA chaperone activity and for the degradation of pulse-labeled misfolded polypeptides. (a) Schematic diagram of the proposed model for the SSA hsp70 chaperone inactivation. (b) The bar graph shows Cdc28-normalized ubiquitylation levels in SSA1 and ssa1-45 cells (ssa2-4Δ) measured before and after shifting cells from 25°C to 37°C for 40 min. The ubiquitylation levels were quantified by dot blot assay from four replicates and shown with standard errors. (c) The increased ubiquitylation levels in ssa1-45 HUL5 or ssa1-45 hul5Δ cells (two independent strains) were compared using five replicates for each assessed strain as described for (b). The p values were determined by Student’s t-test. (d) Wild-type (BY4741) and hul5Δ cells were subjected to 35S-pulse labeling (5 min) followed by a 3 h chase. The graph shows the averaged percentage (with standard errors) of pulse-labeled proteins that were degraded at each time point from cells incubated at 25°C or 38°C from three replicates. (e) Histograms of averaged ubiquitylation signals measured by dot blots in unstressed (no HS; light grey) and heat-shocked (HS; dark grey) cells in three replicates are shown with standard deviations. Cells were grown at 25°C and pre-treated or not for 15 min with 100 μg/ml cycloheximide (CHX) prior to heat-shock (15 min at 45°C). Student’s t-test was used to assess the significance of the ubiquitylation level differences between the indicated strains.