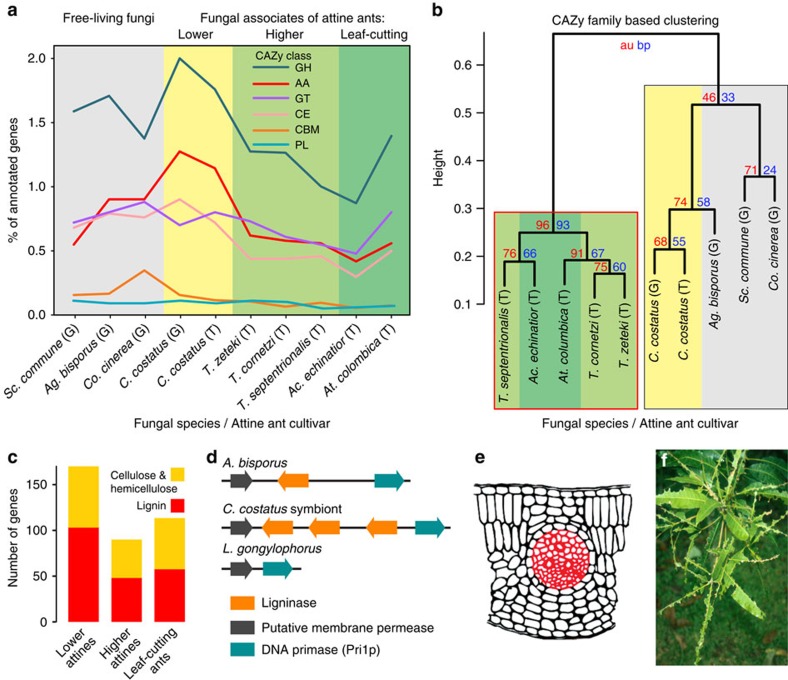
Figure 3. Evolutionary changes in carbohydrate-degrading potential of attine fungal cultivars.
The carbohydrate-degrading potential of attine cultivars is compared with free-living outgroups. Comparisons are based on genomic (G) or transcriptomic (T) gene counts, both of which were obtained independently for C. costatus. (a) Percentage of all annotated genes in the main CAZy classes: AA, auxiliary activities; CBM, carbohydrate-binding modules; CE, carbohydrate esterases; GH, glycoside hydrolases; GT, glycosyltransferases; PL, polysaccharide lyases. Background colours as in Fig. 1; free-living fungi grey. (b) Hierarchical clustering based on genome-wide proportion of CAZy genes per family with approximately unbiased P-values (au, red), standard bootstrap percentages (bp, blue) and the significantly distinct cluster of higher attine cultivars framed in red. (c) Substrate-specific changes in the number of cultivar CAZyme genes for the two major plant cell wall-degrading enzyme classes (hemi) celluloses and lignins. The C. costatus cultivar gene numbers (‘Lower attines') are transcriptome-based to be comparable to those of the higher attine and leaf-cutting ant cultivars. (d) The loss of genes encoding a fungal ligninase domain in higher attine cultivars. Free-living A. bisporus has one copy of the ligninase gene (orange) surrounded by up- and downstream genes (grey and blue). The C. costatus cultivar maintains the gene order but has three tandemly arrayed copies, whereas ligninase genes have been lost in L. gongylophorus. (e) Schematic cross-section of a leaf fragment, with the lignin-rich midrib shown in red. (f) Image illustrating how Panamanian Atta workers avoid the lignin-rich midribs when defoliating understory trees (Photo J.J.B.).