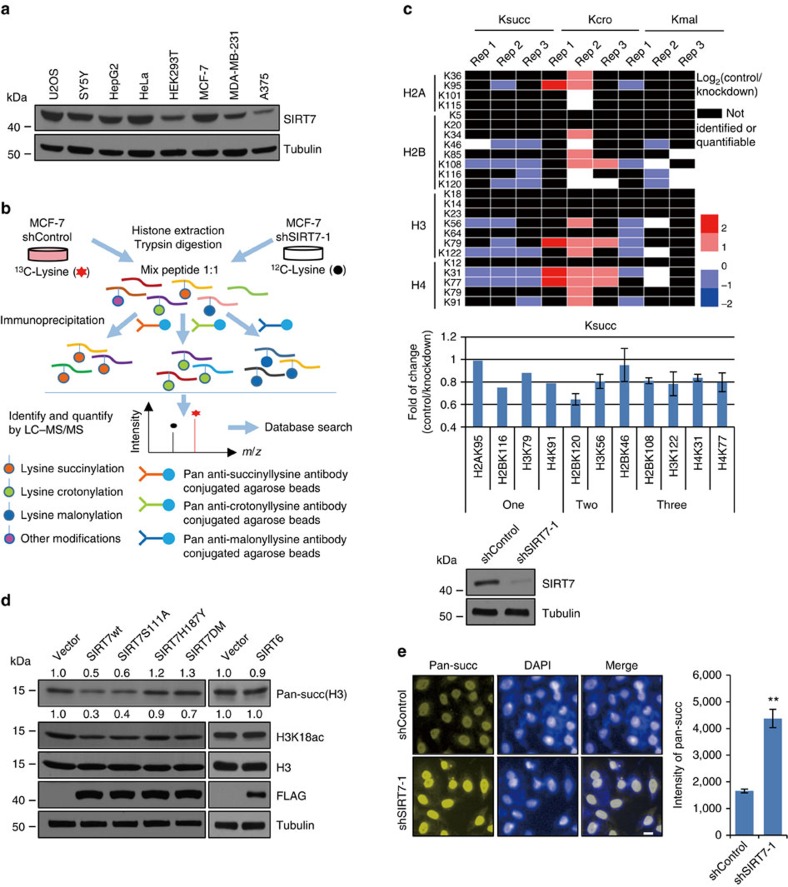
Figure 1. SIRT7 regulates histone succinylation.
(a) Western blotting analysis of SIRT7 expression in different cell lines. (b) The workflow of the integrated SILAC labelling, affinity enrichment and mass spectrometry-based quantitative proteomics to quantify dynamic changes of histone lysine crotonylation (Kcro), succinylation (Ksucc) and malonylation (Kmal) in control or SIRT7-depleted MCF-7 cells. (c) Heatmap of the changes in the levels of Kcro, Ksucc and Kmal of histones detected by SILAC labelling and mass spectrometry-based quantitative proteomics. All experiments were carried out in triplicate and the results are presented as base 2 logarithmic value of the ratio of control/KD. Results of detectable succinylation in one, two or three replicates are also presented as histogram. The efficiency of SIRT7 KD in MCF-7 cells was monitored by western blotting. (d) HEK293T cells were transfected with wild-type SIRT7, SIRT7 mutants or SIRT6. Histones were extracted and pan-succinylation of H3 and H3K18ac were analysed by western blotting. The bands were quantified with ImageJ software. The numbers indicate the relative levels of the indicated modifications. Whole-cell lysate was prepared for monitoring the efficiency of overexpression of SIRT7 and SIRT6 by western blotting. (e) High-throughput microscopic analysis of the mean relative fluorescence intensity of pan-succinylation in control or SIRT7-depleted MCF-7 cells. Scale bar, 10 μm. Each bar represents the mean±s.d. for triplicate experiments. **P<0.01 (two-tailed unpaired Student's t-test).