Fig. 3.
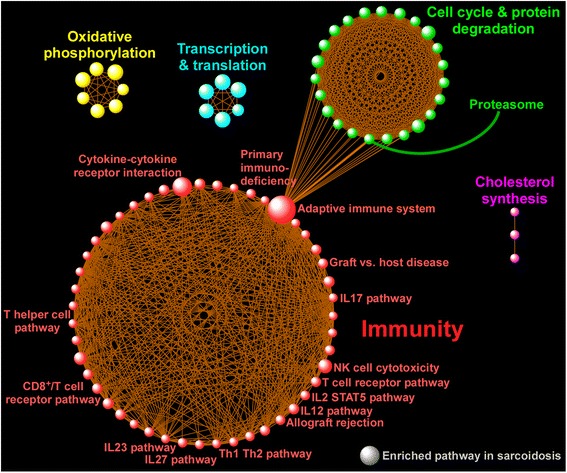
Integrative network analysis of enriched pathways in sarcoidosis as identified using GSEA. A gene set was considered enriched for a given phenotype if most of its member genes were upregulated in that condition. 83 gene sets were upregulated in patients with sarcoidosis whereas very few pathways were enriched in the controls (not shown). In the figure, each sphere designates an enriched pathway in sarcoidosis and the size of each sphere (i.e., gene set) is proportional to the number of its gene members. Only a selected number of pathways have been labeled due to space restriction, but full list is available at Additional file 3: Table S2. Since pathways can share common genes, connectivity lines have been drawn to link these inter-pathway relationships and define the topology of the enrichment network by identifying larger aggregates of functionally associated pathways known as modules. The most prominent module associated with sarcoidosis was “immunity” and incorporated T-cell signaling (T helper and CD8), graft vs. host disease, IL12, IL23, and IL17 pathways among others, but also note the presence of other modules such as “oxidative phosphorylation” and “cell cycle/protein degradation”. Importantly, some pathways from different modules were also linked together, such as the inter-modular connectivity of “adaptive immune system” with “proteasome” as shown in the figure. This relationship indicates that these two gene sets share common functional characteristics and implicates proteasomal processes as potential modulators of adaptive immunity in sarcoidosis
