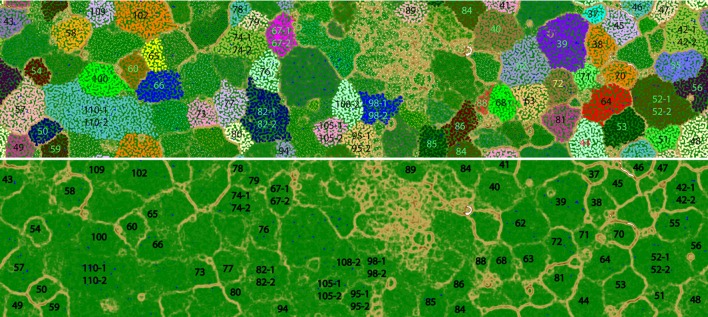
FIGURE 1.
Tetra-nucleotide ESOM binning map of Guaymas Basin plume metagenomic assembly. (Top) Each point on the map represents a contig (>5 kb) or contig fragment generated in silico (5–10 kb). All identified bins are uniquely color coded as indicated. The ESOM map displayed is tiled and torroidal (that is, continuous from top to bottom and left to right). Note that in a few cases (Bin42, Bin52, Bin67, Bin74, Bin82, Bin95, Bin98, Bin105, Bin108, Bin110) multiple closely related bins fall within one large cluster on this map. These bins were delineated based on differential coverage plots. (Bottom) the same ESOM image, without data points shown, highlighting the topographic structure used to delineate bins. Higher “elevations” (brown, white) indicate large tetranucleotide frequency distances between data points, representing boundaries between genomic bins. Lower elevations (green, blue”) indicate small tetranucleotide frequency distances between data points, representing the continuity of tetranucleotide frequency space within genomes.