Abstract
A urease-negative, fusiform, novel bacterium named Helicobacter saguini was isolated from the intestines and feces of cotton-top tamarins (CTTs) with chronic colitis. Helicobacter sp. was detected in 69% of feces or intestinal samples from 116 CTTs. The draft genome sequence, obtained by Illumina MiSeq sequencing, for H. saguini isolate MIT 97-6194-5, consisting of ∼2.9 Mb with a G+C content of 35% and 2,704 genes, was annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline. H. saguini contains homologous genes of known virulence factors found in other enterohepatic helicobacter species (EHS) and H. pylori. These include flagellin, γ-glutamyl transpeptidase (ggt), collagenase, the secreted serine protease htrA, and components of a type VI secretion system, but the genome does not harbor genes for cytolethal distending toxin (cdt). H. saguini MIT 97-6194-5 induced significant levels of interleukin-8 (IL-8) in HT-29 cell culture supernatants by 4 h, which increased through 24 h. mRNAs for the proinflammatory cytokines IL-1β, tumor necrosis factor alpha (TNF-α), IL-10, and IL-6 and the chemokine CXCL1 were upregulated in cocultured HT-29 cells at 4 h compared to levels in control cells. At 3 months postinfection, all H. saguini-monoassociated gnotobiotic C57BL/129 IL-10−/− mice were colonized and had seroconverted to H. saguini antigen with a significant Th1-associated increase in IgG2c (P < 0.0001). H. saguini induced a significant typhlocolitis, associated epithelial defects, mucosa-associated lymphoid tissue (MALT) hyperplasia, and dysplasia. Inflammatory cytokines IL-22, IL-17a, IL-1β, gamma interferon (IFN-γ), and TNF-α, as well as inducible nitric oxide synthase (iNOS) were significantly upregulated in the cecal tissues of infected mice. The expression of the DNA damage response molecule γ-H2AX was significantly higher in the ceca of H. saguini-infected gnotobiotic mice than in the controls. This model using a nonhuman primate Helicobacter sp. can be used to study the pathogenic potential of EHS isolated from primates with naturally occurring inflammatory bowel disease (IBD) and colon cancer.
INTRODUCTION
Cotton-top tamarins (CTTs) are New World primates indigenous to the rain forests of Colombia and were imported into the United States for biomedical research beginning in the 1960s, until their classification as endangered species in 1976 (1). Approximately 50% of colony-maintained tamarins develop idiopathic chronic colitis, with 20 to 40% of cases evolving into colonic adenocarcinomas (2). The clinical and histopathological manifestations of colitis in CTTs resemble human inflammatory bowel disease (IBD), particularly ulcerative colitis (UC), making the CTT an attractive animal model of naturally occurring IBD. The etiology of colitis in CTTs remains unknown but has been speculated to be caused by genetic predispositions and/or conditions related to captivity, such as husbandry, the environment (temperature, humidity, and sanitation), abnormal diet, stress, and infectious agents (Escherichia coli, Campylobacter spp., and Helicobacter spp.) (3–5).
In 1999, we detected and isolated a urease-negative, fusiform organism, for which we are proposing the name Helicobacter saguini, from the intestines and feces of CTTs with chronic colitis. Phylogenetic analysis by 16S rRNA gene sequencing classified this isolate as a novel enterohepatic helicobacter species (EHS) (5).
EHS are associated with the development of IBD in immunocompromised mice and have been identified in humans with diarrhea and in some individuals with IBD (6, 7). H. hepaticus, H. bilis, and other EHS infections in immunocompromised mice cause chronic typhlocolitis, high-grade dysplasia, and progression to colitis-associated carcinoma (CAC) (8–11). Novel Helicobacter spp., including H. macacae, have been cultured from idiopathic colitis and colon adenocarcinomas in rhesus monkeys (12–14). H. cinaedi and H. fennelliae were first isolated from homosexual men (presumably immunocompromised by HIV infection) with proctitis (15). Experimentally, H. cinaedi and H. fennelliae infection in pigtail macaques induced diarrhea and inflammation of the lower bowel (16). Additionally, EHS prevalence in human UC patients was shown to be significantly greater than that in healthy individuals; however, the identification and isolation of a Helicobacter sp. implicated in the pathogenesis of human IBD have remained elusive (17).
We hypothesized that H. saguini has a pathogenic potential similar to that of other EHS and could be associated with colitis and colon cancer in CTTs. However, the endangered status and predisposition toward colitis in captivity have precluded direct study of the causal relationship between H. saguini and IBD in CTTs. As an alternative, we used genome analysis, in vitro assays, and an interleukin-10 knockout (IL-10−/−) mouse infection model to investigate the pathogenicity of H. saguini. We report herein that H. saguini harbors putative virulence factors and elicits proinflammatory responses in vitro and in vivo, thus demonstrating that H. saguini has the pathogenic potential to induce IBD in CTTs. This finding adds additional credence to the view that EHS may play a role in inducing IBD in humans (6, 17).
MATERIALS AND METHODS
Fecal and colonic biopsy samples.
Thirty colonic biopsy samples, 88 rectal swabs, and 29 fecal samples were collected from 116 CTTs which were selected from a colony in which colitis was endemic. These samples were collected over a period of 5 years (Table 1).
TABLE 1.
Helicobacter sp. prevalence in a cotton-top tamarin colony
| Helicobacter detection method | No. (%) of positive samples or animals |
|||
|---|---|---|---|---|
| Anal swab (n = 88) | Fecal sample (n = 29) | Biopsy specimen (n = 30) | Total animal population (n = 116) | |
| PCRa | 57/88 (64.8) | 22/29 (75.9) | 24/30 (80) | 80/116 (69) |
| Culture | 2/18 (11.1) | 1/9 (11.1) | 8/30 (26.7) | 11/57 (19.3) |
Select animals had more than one PCR analysis.
Helicobacter sp. PCR.
A High Pure PCR template preparation kit (Roche Molecular Biochemicals, Indianapolis, IN) was used for extraction of DNA from the bacterial isolates and the tissue samples; a QIAamp DNA Stool minikit was used for rectal swabs and fecal sample DNA extraction according to the manufacturer's directions (Qiagen, Valencia, CA). Helicobacter genus-specific primers C97 (5′-GCT ATG ACG GGT ATCC) and C05 (5′-ACT TCA CCC CAG TCG CTG) were used to amplify a 1.2-kb PCR product from the 16S rRNA gene (18). The 1,200-bp PCR products were sequenced using previously described techniques (18). A TOPO-TA Cloning kit was used to clone and sequence PCR products with mixed signals in the direct sequencing reactions (Life Technologies, Grand Island, NY) according to the manufacturer's instructions.
Culture and characterization of H. saguini.
Eighteen rectal swabs, 9 fecal samples, and 30 biopsy samples were also subjected to microaerobic culture. Rectal swabs and fecal samples were diluted in brucella broth with 20% glycerol; biopsy samples were homogenized, and an aliquot of each slurry was placed on cefoperazone-vancomycin-amphotericin B (CVA) plates and passed through a 0.65-μm-pore-size syringe filter onto a Trypticase soy agar plate with 5% sheep blood (Remel Laboratories, Lenexa, KS). The plates were incubated at 37°C under microaerobic conditions in a vented jar containing N2, H2, and CO2 (80:10:10) and evaluated for bacterial growth every 2 to 3 days for 3 weeks. Detailed biochemical characterization analysis was performed on eight individual isolates using a RapID NH System (Remel Laboratories, Lenexa, KS) and API Campy kits (bioMérieux, Boston, MA). Urease, catalase, and oxidase production, sensitivity to nalidixic acid and cephalothin, and growth in the presence of 1% glycine were determined as previously described by our laboratory (19). A disc assay was used for indoxyl acetate hydrolysis (20). Suspected bacterial growth was identified as Helicobacter spp. on the basis of colony morphology, phase microscopy, Gram staining, biochemical testing, Helicobacter-specific PCR, and 16S rRNA and 23S rRNA gene sequencing (21).
Electron microscopy.
H. saguini isolate MIT 97-6194-5 was examined by electron microscopy. Cells grown on blood agar plates were centrifuged and gently suspended in 10 mM Tris-HCl buffer (pH 7.4) at a concentration of about 108 cells per ml. Samples were negatively stained with 1% (wt/vol) phosphotungstic acid (pH 6.5) for 20 to 30 s. The specimens were examined with a JEOL model JEM-1200EX transmission electron microscope operating at 100 kV.
H. saguini whole-genome sequence analysis.
Genomic DNA from H. saguini MIT 97-6194-5 was sequenced using Illumina MiSeq sequencing technology as described previously (22). The 250-bp paired-end sequencing reads generated by MiSeq were assembled into contigs using Velvet (23). Sequences were annotated using the NCBI Prokaryotic Genomes Automatic Annotation Pipeline (24). The whole genomic distance between H. saguini MIT 97-9694-5 and its nearest neighbor, H. jaachi MIT 09-6949, was calculated using the Genome-to-Genome Distance Calculator (GGDC [http://ggdc.dsmz.de]) (25).
Interactions of H. saguini with human intestinal epithelial cell line HT-29.
HT-29 cells (colorectal adenocarcinoma) were obtained from the American Type Culture Collection (ATCC) and maintained in Dulbecco's modified Eagle's medium (DMEM) (Sigma-Aldrich, St. Louis, MO) containing 10% fetal bovine serum (FBS) (Sigma) and 1% antibiotic-antimycotic (Thermo Fisher Scientific, Grand Island, NY) at 37°C with 5% CO2. HT-29 cells were seeded into six-well cell culture plates and incubated at 37°C in a humidified incubator with 5% CO2 for 48 h. H. saguini cells were passaged on blood agar under microaerobic conditions and collected after 48 h, which corresponded to the mid-exponential phase of the bacterial growth. Viable organisms were predominantly fusiform and demonstrated rapid movement when observed under phase-contrast microscopy. Gram staining was used to ensure the purity of the bacterial preparation. HT-29 cells (2.5 × 106 cells/well) were inoculated with H. saguini at multiplicities of infection (MOIs) of 1:100 and 1:25 in 2 ml of fresh culture medium containing 1% FBS without antibiotics. The plates were centrifuged at 200 × g to facilitate bacterial cell adhesion and then incubated under 5% CO2. After 4 and 24 h of incubation, cell supernatants were collected after centrifugation for determination of IL-8 concentration by enzyme-linked immunosorbent assay (ELISA; Qiagen). HT-29 cells were washed once with phosphate-buffered saline (PBS) and collected in TRIzol reagent for total RNA extraction.
Experimental infection of C57BL/129 IL-10−/− mice with H. saguini MIT 97-6194-5.
Forty specific-pathogen-free (SPF) and 14 germfree (GF) C57BL/6 (B6.129P2-IL-10tm1Cgn [IL-10−/−]) mice, aged 6 to 8 weeks old, were used in the infection study, with similar numbers of male and female mice. All mice were from a breeding colony maintained at the Massachusetts Institute of Technology (MIT). SPF mice were maintained free of known murine viral pathogens, Salmonella spp., Citrobacter rodentium, ecto- and endoparasites, and known Helicobacter spp. in an AAALAC International-accredited facility under barrier conditions. Animals were housed in microisolater, solid-bottomed polycarbonate cages on hardwood bedding, fed a commercial pelleted diet (Prolab RMH 3000; PMI Nutrition International), and administered water ad libitum.
Germfree and monoassociated mice were housed in sterile isolators on autoclaved hardwood bedding in sterile, solid-bottomed polycarbonate cages and fed autoclavable Mouse Breeder Diet 5021 (PMI Nutrition International) and sterile water ad libitum. Isolators were surveyed biweekly and confirmed negative for microbial contaminants. The protocol was approved by the Committee on Animal Care of the Massachusetts Institute of Technology.
H. saguini MIT 97-6194-4 was grown under microaerobic conditions at 37°C on 5% sheep blood agar plates for 2 days. Bacteria were collected and resuspended in brucella broth with 20% glycerol and adjusted to a bacterial concentration of 1 optical density unit at 600 nm (OD600)/ml. Half of the mice received 0.2 ml of fresh inoculum by gastric gavage every other day for three doses, and the other half were sham dosed with broth only. Colonization with H. saguini was confirmed 2 weeks postinoculation by PCR analysis of feces using Helicobacter genus-specific primers (18). SPF mice were necropsied at 4 weeks postinfection (wpi), and monoassociated mice were necropsied at 3 months postinfection (p.i.). Serum and feces from SPF and monoassociated mice were collected, followed by liver, stomach, cecum, and colon, for culture, quantitative PCR, RNA isolation, and histology.
Detection of serum antibody responses to H. saguini by ELISA.
Sera were collected at necropsy, and total IgG along with Th1-associated IgG2c and Th2-associated IgG1 responses to antigens of H. saguini were measured by ELISA (26). Sonicated antigen from H. saguini MIT 97-6194-5 was coated on Immulon II plates (Thermo Labsystems, Franklin, MA) at a concentration of 1 μg/ml (total IgG) or 10 μg/ml (subclass isotypes), and sera were diluted 1:100. Biotinylated secondary antibodies included goat anti-mouse IgG (Southern Biotechnology Associates, Pittsburgh, PA) and monoclonal anti-mouse antibodies (BD PharMingen, San Jose, CA) for detecting IgG1 and IgG2c. Incubation with ExtrAvidin peroxidase (Sigma, St. Louis, MO) was followed by avidin-biotin-peroxidase complex (ABTS) substrate (Kirkegaard and Perry Laboratories, Gaithersburg, MD) for color development. Optical density (OD) development at a wavelength (λ) of 405 nm was recorded by an Epoch spectrophotometer (BioTek, Winooski, VT).
Histological evaluation.
Formalin-fixed tissues were routinely processed, embedded in paraffin, cut at 4 μm, and stained with hematoxylin and eosin (H&E). Large bowel lesions were scored on the basis of size and frequency of hyperplastic and inflammatory lesions on a scale of 0 to 4 with ascending severity (0, none; 1, minimal; 2, mild; 3, moderate; and 4, severe). Epithelial dysplasia and neoplasia were graded using a scale of 0 to 4, as follows: 0, normal; 1, mild dysplastic changes; 2, moderate to severe dysplasia; 3, gastrointestinal intraepithelial neoplasia (GIN); and 4, invasive carcinoma as previously described (8, 9).
Fluorescent in situ hybridization (FISH) for Helicobacter species.
Paraffin sections of colons were deparaffinized and rehydrated. Helicobacter genus-specific probes, HEL274 and HEL717, labeled with Cy3 were used (Integrated DNA Technologies) (27). Hybridization buffer (0.9 M NaCl, 100 mM Tris-HCl, 0.1% SDS, 30% formamide) with 5 ng of probe ml−1 was preheated for 10 min at 74.5°C; 80 μl of this solution was added to each slide. Slides were covered in Parafilm and placed in a dark humidification chamber overnight at 48°C. After incubation, slides were rinsed in double-distilled water and serially washed in prewarmed rinsing buffers for 15 min each (buffer 1 consisting of 0.9 M NaCl, 100 mM Tris-HCl, and 0.01% SDS; buffer 2 consisting of 0.9 M NaCl, and 100 mM Tris-HCl). Slides were air dried, mounted in Vectashield with 4′,6′-diamidino-2-phenylindole (DAPI; Vector Laboratories), and examined under a Zeiss Axioskop 2 fluorescence microscope. Tissues were considered positive for Helicobacter spp. if fluorescent spiral organisms were observed under the rhodamine filter (21).
Immunohistochemical analysis of γ-H2AX.
The paraffin-embedded mouse cecum tissues (5 μm thick) were prepared, and after the tissue sections were deparaffinized and rehydrated in graded ethanol concentrations, the slides were immersed in low-pH target retrieval solution (Dako, Carpinteria, CA, USA) in a 95°C water bath for 20 min. The slides were washed in Tris-buffered saline (TBS) and blocked with 3% bovine serum albumin (BSA) in TBS overnight at 4°C. The slides were incubated with monoclonal rabbit anti-mouse γ-H2AX antibody (1:200 dilution; Cell Signaling, Danvers, MA, USA) for 2 h, followed by incubation for 90 min with Alexa Fluor 488-conjugated anti-rabbit F(ab′)2 fragment (Cell Signaling). To stain the nuclei, the slides were mounted using 10 μl of Prolong Gold antifade reagent with DAPI (Cell Signaling). The tissue sections were observed using a Zeiss Axioskop 2 Plus microscope (Zeiss, Germany), and cells possessing one or more γ-H2AX+ foci were counted as positive. The results are presented as the average percentage of positive epithelial cells observed in 8 to 10 high-power images.
Cytokine mRNA expression profiles in HT-29 cells and the cecum and colon of monoassociated C57BL/129 IL-10−/− mice.
RNA was extracted from cells and mouse cecum tissues using TRIzol reagent (Invitrogen, Carlsbad, CA). Total RNA (2 μg) was converted into cDNA using a High Capacity cDNA Archive kit according to the manufacturer's protocol (Applied Biosystems). cDNA levels for tumor necrosis factor alpha (TNF-α), gamma interferon (IFN-γ), IL-1β, IL-4, IL-6, IL-17α, IL-22, IL-23α, CXCL1, and inducible nitric oxide synthase (iNOS) mRNAs were measured by quantitative PCR using commercial primers and probes for each cytokine. Briefly, duplicate 20-μl reaction mixtures contained 5 μl of cDNA, 1 μl of a commercial 20× primer-probe solution, 10 μl of 2× master mix (all Applied Biosystems), and 4 μl of double-distilled H2O. Relative expression of mRNA from infected and control tissues was calculated using the comparative threshold cycle (CT) method with RNA input standardized between samples by expression levels of the endogenous reference gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH). Results from duplicate samples were plotted as fold changes between cells or tissues from infected and uninfected controls.
Statistical analysis.
Cecal and colonic lesion scores were analyzed using a Mann-Whitney U nonparametric test for ordinal data; levels of cytokine mRNA expression and serology results evaluated were compared by Student's t test. Statistical analysis was performed using GraphPad Prism, version 5.0 (GraphPad Software, Inc., La Jolla, CA). Results were considered significant at a P value of <0.05.
Nucleotide sequence accession numbers.
The draft genome sequence of H. saguini has been submitted to GenBank under accession number JRMP00000000. The 16S rRNA sequence of the type strain of H. saguini MIT 97-6194-5 has been deposited in GenBank under accession number AF107494.1.
RESULTS
Helicobacter saguini is highly prevalent in cotton-top tamarins.
H. saguini was originally isolated from colony-maintained CTTs by our laboratory in 1999 (5), where 53% of colon biopsy samples were found to be positive for Helicobacter spp. by genus-specific Helicobacter sp. PCR. We further assessed the prevalence of Helicobacter infection in CTTs by surveying samples previously collected in 2001 and 2006 from animals living in the same colony in which colitis was endemic; 65% of anal swabs, 76% of feces, and 80% of colon biopsy samples were PCR positive for Helicobacter spp. In total, Helicobacter spp. were detected in 69% of the animals (Table 1). Representative PCR products from two anal swab samples, nine biopsy samples, and two fecal samples had 16S rRNA sequences identical to those of H. saguini strain MIT-97-6194-5. Eleven Helicobacter isolates successfully cultured from 57 samples had 16S rRNA sequences identical to those of H. saguini strain MIT 97-6194-5 (Table 1). PCR products from four biopsy samples had mixed sequences and were cloned into a pCR2.1-TOPO TA vector; eight colonies from each PCR-positive biopsy sample were sequenced. In addition to H. saguini, two other Helicobacter spp. were present. They had 97% sequence homologies with Helicobacter spp. previously identified in the feces of a rhesus macaque (MIT 01-3238) and the colon of a baboon (MIT 03-7674c), respectively.
Twenty-three CTTs had both anal swabs and biopsy samples submitted for testing in 2001. Overall, 20/23 (86.9%) of the animals were PCR positive. The Helicobacter PCR results for both types of samples correlated very well (overall, 21/23, or 91.3%, with the same result; both positive, 18/23, or 78.3%; both negative, 3/23, or 13%); only 2/23 (8.7%) had different results between sample types. These results suggest that anal swabs can be used as an alternative to the invasive biopsy sampling method for H. saguini detection. Feces from 4 of the 23 CTTs were retested on 2006; all remained Helicobacter positive, indicating a chronic, persistent Helicobacter infection in these animals.
Biochemical analysis.
The biochemical characteristics of eight H. saguini isolates (four from original isolates in 1999 and four from the current study) were compared with those of other Helicobacter species (Table 2). All isolates were oxidase and catalase positive and urease negative. The isolates did not reduce nitrate to nitrite, did not hydrolyze alkaline phosphatase, and were not able to hydrolyze indoxyl acetate. Five of the eight isolates had γ-glutamyl transpeptidase activity, and all were resistant to nalidixic acid and cephalothin. The organism grew in 1% glycine and at 37 and 42°C, but not at 25°C. The type strain of H. saguini MIT 97-6194-5 has been deposited in the BCCM/LMG Bacteria Collection as LMG 28611.
TABLE 2.
Comparison of biochemical tests for Helicobacter saguini and other Helicobacter spp.a
| Taxon | Catalase | Oxidase | NO3 | Urease | IAH | GGT | PO4 | Growth at 42°C | Growth with 1% glycine | Drug resistancec |
DNA G+C content (mol%) | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NA | CE | |||||||||||
| Helicobacter saguinib | + | + | − | − | − | + | − | + | + | R | R | 34.6 |
| Helicobacter jaachi | + | + | − | + | + | − | − | + | + | S | R | 41 |
| “Helicobacter callitrichis” | + | + | − | − | − | − | + | + | + | S | R | 39.6 |
| Helicobacter macacae | + | − | − | − | − | − | − | + | + | R | R | 40.6 |
| Helicobacter trogontum | + | + | + | + | − | + | − | + | ND | R | R | 34 |
| Helicobacter bilis | + | + | + | + | − | + | − | + | + | R | R | ND |
| Helicobacter marmotae | + | + | − | + | − | − | + | − | + | R | R | ND |
| Helicobacter canis | − | + | − | − | + | + | + | + | − | S | I | 45 |
| Helicobacter hepaticus | + | + | + | + | + | − | − | − | + | R | R | 35.9 |
| Helicobacter pylori | + | + | − | + | − | + | + | − | − | R | S | 35–37 |
NO3, nitrate reduction; IAH, indoxyl acetate hydrolysis; GGT, γ-glutamyl transpeptidase; PO4, alkaline phosphatase hydrolysis.
Data were modified from our 1999 paper (5) and represent 4 strains from the original study and 4 strains from 2006 isolates. In all cases except for GGT, the results represent 8/8 strains. For GGT, the result represents 5/8 strains.
NA, nalidixic acid; CE, cephalothin; R, resistant; S, susceptible.
Electron microscopy.
By electron microscopy, cells of H. saguini had a fusiform appearance and measured approximately 0.5 by 4 to 5 μm (Figure 1). The cells possessed periplasmic fibers and 6 to 12 bipolar, sheathed flagella.
FIG 1.
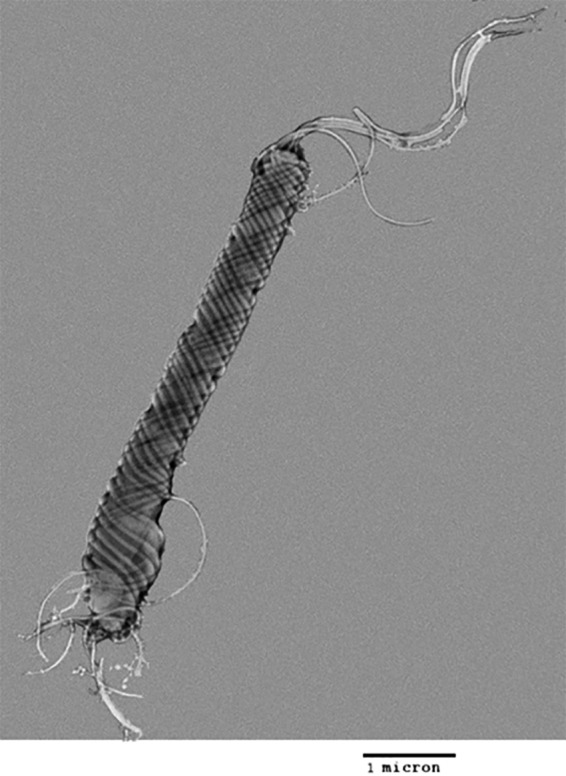
Transmission electron micrograph of negatively stained H. saguini MIT 97-6194-5.
Phylogenetic analysis.
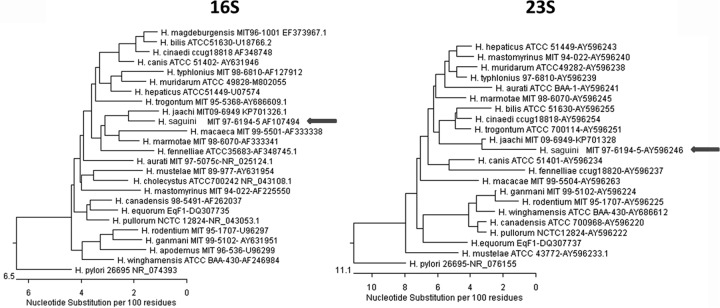
Full 16S rRNA genes from six H. saguini strains were sequenced. They shared over 99% sequence similarity with each other. The most closely related species was H. jaachi, isolated from common marmosets (97% similarity with the type strain 16S rRNA sequence) (Fig. 2). The 23S rRNA gene from H. saguini MIT 97-6194-5 was also compared with the 23S rRNA sequences of other Helicobacter species. The closest related species was H. jaachi, which also had a sequence similarity of 97% to the H. saguini type strain 23S sequence (Fig. 2). Finally, the hsp60 gene of H. saguini obtained from the whole-genome sequence of MIT 97-6194-5 was compared with the 600-bp conserved region of the hsp60 gene in other Helicobacter spp. The most closely related species was H. canis (79% sequence identity to that of the H. saguini type strain hsp60 sequence).
FIG 2.
Phylogenetic analysis of 16S rRNA and 23S rRNA gene sequences. Neighbor-joining trees were based on the comparison of genes from different Helicobacter species. Arrows indicate H. saguini.
Draft genome of H. saguini.
The H. saguini type strain MIT 97-6194-5 genome measures 2.92 Mb in size with a G+C content of 34.6%, and it contains 2,704 genes encoding 2,294 proteins, 5 rRNAs, 39 tRNAs, 1 other RNA, and 365 pseudogenes. Homologous genes of previously described virulence factors from Helicobacter spp. and related species were identified in the annotated genome, including flagellum components, flavodoxin fldA, γ-glutamyl transpeptidase (ggt), the secreted serine protease htrA, and the type VI secretion component vgfG. Unlike other selected EHS, H. saguini does not harbor genes for cytolethal distending toxin (cdt). However, similar to other EHS, H. saguini also lacks vacA or cagA (Table 3).
TABLE 3.
Profile of draft genomes of H. saguini, H. pylori, and H. hepaticus
| Characteristic | H. pylori 26695 | H. saguini LMG 28611 | H. hepaticus ATCC 51449 |
|---|---|---|---|
| Genome size (Mb) | 1.67 | 2.9 | 1.8 |
| G+C content (%) | 39 | 34.6 | 35.9 |
| No. of genes | 1,590 | 2,704 | 1,875 |
| Selected virulence genesa | cagA, vacA, ggt, nap, T4SS, htrA, ure | ggt, T6SS, htrA | T6SS, ure, cdt |
vacA, vacuolating cytotoxin A; cagA, cytotoxin-associated gene; ggt, γ-glutamyl transpeptidase; nap, neutrophil-activating protein; T4(6)SS, type IV(VI) secretion system; htrA, high-temperature requirement A protein-secreted serine protease; ure, urease.
In silico comparison of the H. saguini MIT 97-6194-5 genome sequence with that of H. jaachi MIT 09-6949 (GenBank accession number JRPR00000000), its closest phylogenetic neighbor, revealed 12.90% ± 2.98%, 23.30% ± 2.38%, and 13.30% ± 2.55% similarity, using the three formulas offered by the Genome-to-Genome Distance Calculator (GGDC) (25), version 2.0. These data unambiguously demonstrate that both strains represent distinct species, thus confirming the 6.38% difference in mol% G+C content between the two genome sequences (28).
H. saguini induces proinflammatory cytokines and chemokines in the human intestinal epithelial cell line HT-29.
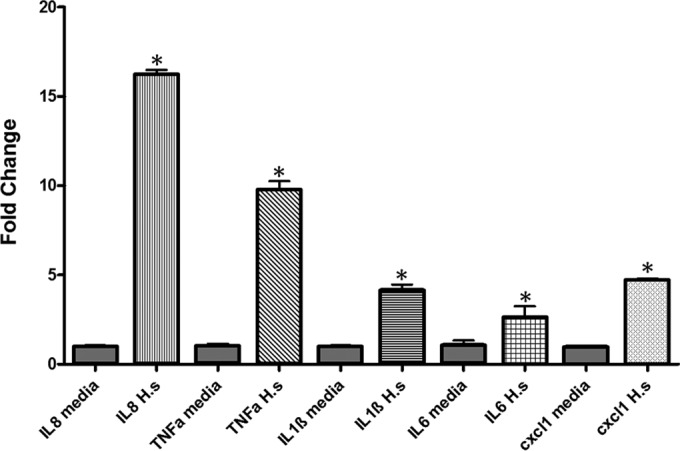
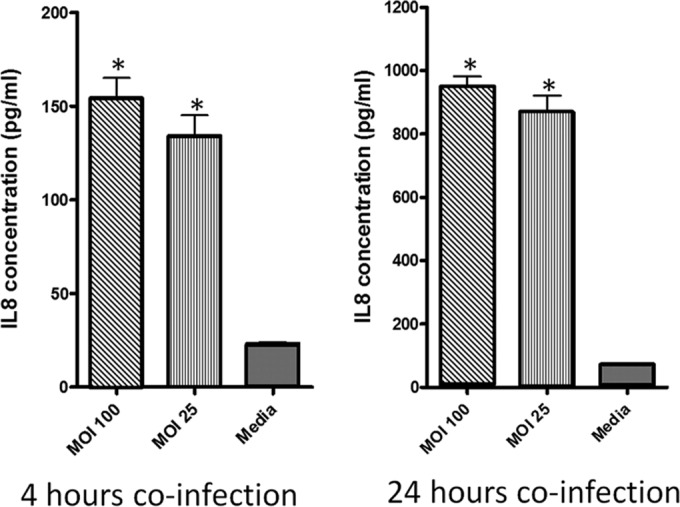
Human intestinal epithelial cells (HT-29) were cocultured with H. saguini (MIT 09-6949-5) at an MOI of 25 or 100 for 4 and 24 h. Inflammatory cytokine gene expression of the HT-29 cells and secreted IL-8 in cell culture supernatant were quantified to determine if H. saguini is capable of inducing a proinflammatory response. H. saguini stimulated IL-8 production in HT-29 cells. IL-8 concentration in the supernatant of H. saguini-infected cells was significantly higher than that in the medium control at an MOI of 1:100 (P < 0.001). IL-8 levels were 150 pg/ml at 4 h and 900 pg/ml at 24 h in HT-29 cells infected with H. saguini (Fig. 3). mRNA expression levels of IL-8, TNF-α, IL-1β, IL-6, and CXCL1 were significantly increased (P < 0.001) compared to levels in the medium controls 4 h after infection at an MOI of 1:100 (Fig. 4).
FIG 3.
H. saguini stimulated IL-8 production by HT-29 cells. HT-29 cells were coincubated with H. saguini at an MOI of 25 or 100 or with medium alone for 4 and 24 h. IL-8 concentrations in the supernatants were measured by ELISA. *, P < 0.001.
FIG 4.
Expression levels of proinflammatory genes in HT-29 colonic cell line in medium alone or coincubated with H. saguini (H.s.) for 4 h at an MOI of 100. Data represent means and standard errors of fold changes in mRNA expression levels. *, P < 0.01.
Experimental H. saguini successfully colonizes germfree C57BL/129 IL-10−/− mice.
Specific-pathogen-free (SPF) and germfree (GF) C57BL/129 IL-10−/− mice were dosed with 2 × 108 CFU of H. saguini by oral gavage every other day three times. H. saguini successfully colonized GF mice. Feces from all monoassociated mice infected with H. saguini were PCR positive for Helicobacter spp. at 2 weeks postinfection (wpi), and sequencing of the 16S rRNA PCR products confirmed the organisms to be H. saguini. Additionally, H. saguini was cultured from gnotobiotic mouse feces. Monoassociated mice euthanized at 3 months postinfection were also positive for Helicobacter spp. by PCR. In contrast, SPF IL-10−/− mice were not colonized by H. saguini, as determined by PCR-negative results for Helicobacter spp. at 2 wpi in fecal samples. None of the SPF mice at necropsy had gross changes at 4 wpi, and they were PCR negative for Helicobacter in colon samples.
FISH.
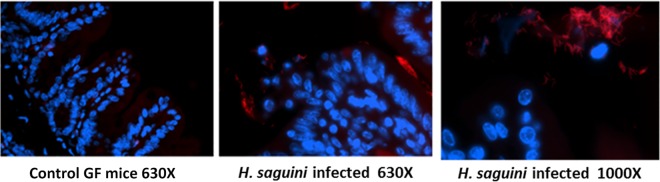
A Helicobacter genus-specific probe was used to identify H. saguini in monoassociated cecal tissues. H. saguini, as demonstrated by fluorescence in situ hybridization (FISH), was present in the lumen and crypts of the colons and ceca of monoassociated mice (Fig. 5).
FIG 5.
Fluorescence in situ hybridization (FISH) using a Helicobacter genus-specific probe in monoassociated mouse cecum. H. saguini cells are labeled in red. DAPI stained the nuclei blue.
H. saguini induces significant typhlocolitis in monoassociated mice.
In monoassociated mice, H. saguini induced typhlocolitis characterized by moderate to severe infiltration of inflammatory cells, including lymphocytes, histiocytes, and neutrophils, into the mucosa and submucosa. Also present were epithelial defects, mucosa-associated lymphoid tissue (MALT) hyperplasia, and dysplasia (Fig. 6A to C). H. saguini-monoassociated mice had significantly higher inflammation, hyperplasia, and dysplasia scores than control mice in both the cecum and colon (P < 0.05). GF control mice did not have overt histologic signs of lower bowel inflammation and had normal crypts and epithelial surfaces in the colon and cecum.
FIG 6.
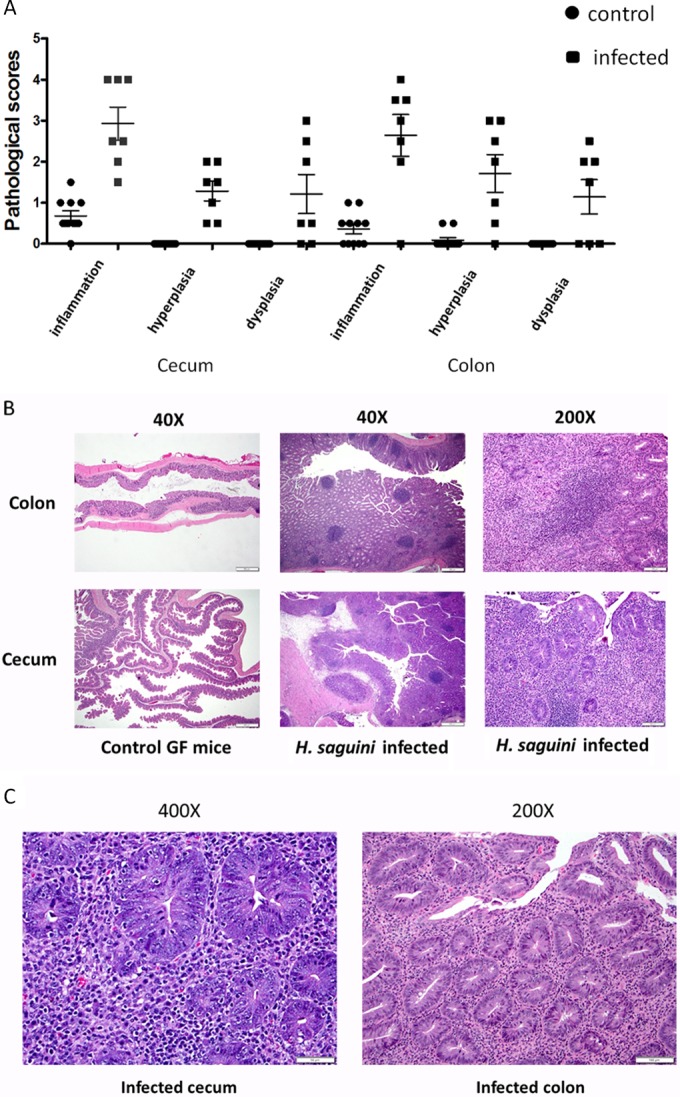
(A) H. saguini-infected monoassociated IL-10−/− mice had significantly higher inflammation, hyperplasia, and dysplasia scores than GF mice in both cecum and colon (P < 0.05). (B and C) H. saguini-induced typhlocolitis characterized by moderate to severe infiltration of inflammatory cells, including lymphocytes, histiocytes, and neutrophils, into the mucosa and submucosa, with epithelial defects, mucosa-associated lymphoid tissue (MALT) hyperplasia, and dysplasia (H&E staining).
H. saguini infection induces H2AX-positive epithelial cells.
Double-stranded breaks (DSBs) are defined as DNA damage in which two complementary strands of double-helix DNA are damaged simultaneously in a location close to each other. It is considered the most dangerous type of DNA damage, and unrepaired DSBs are sufficient for induction of cell death. DSBs can be initiated in response to a variety of stress signals that are encountered during physiological processes. When cells are exposed to ionizing radiation, DNA-damaging chemotherapeutic agents, or chronic infection by Helicobacter spp., DSBs are generated that rapidly result in the phosphorylation of histone H2A variant H2AX. Because phosphorylation of H2AX at Ser139 (γ-H2AX) correlates well with each DSB, it is the most sensitive marker that can be used to examine DNA damage and the subsequent repair of the DNA lesion (29–31).
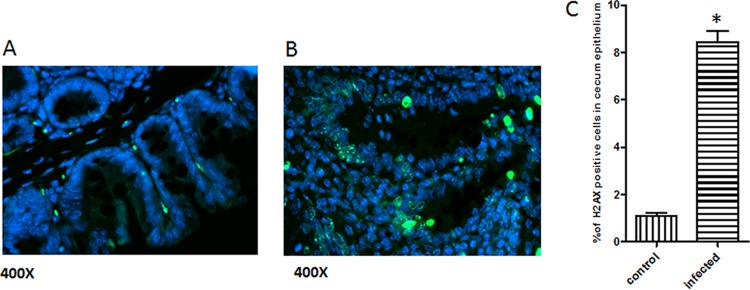
Immunofluorescence staining of γ-H2AX was performed on the ileo-cecal-colic junction of GF and monoassociated mice. In the H. saguini-monoassociated mice, significantly increased numbers of γ-H2AX-positive cells were observed in the intestinal epithelium, particularly within the regions of intestinal hyperplasia and dysplasia; the uninfected tissue had very few positively stained cells (Fig. 7).
FIG 7.
Immunofluorescence staining of gnotobiotic mouse cecum for γ-H2AX (in green). Five mice in each group were examined, with the following results: the uninfected tissue had very few positively stained cells in the cecum (A) and more γ-H2AX-positive cells were observed in the epithelium, particularly within areas of intestinal hyperplasia and dysplasia in the infected cecum (B). (C) The number of γ-H2AX-positive cells in the cecal epithelium of H. saguini-infected mice was statistically higher than that in the control. (*, P < 0.05).
H. saguini has in vivo proinflammatory properties.
Consistent with histological evidence of inflammation, H. saguini induced mRNAs for proinflammatory cytokines and chemokines in the cecal tissue of infected mice compared to levels in control mice (P < 0.001) (Fig. 8). The most significantly increased cytokines were IL-22, IL-17a, IFN-γ, TNF-α, and IL-6; iNOS expression was increased as well; no significant changes were noted in IL-23a and IL-4 mRNA expression levels (data not shown).
FIG 8.
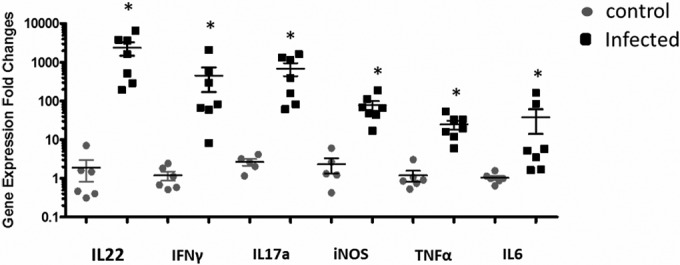
Inflammatory gene expression in cecal tissue at 3 months postinfection. Expression of each inflammatory gene was significantly greater in infected than in control mice (*, P < 0.001). The data are presented as the fold change compared to mean gene expression of GAPDH.
H. saguini induces a systemic immune response.
Serum antibody against H. saguini was measured by ELISA. Total IgG and Th1-associated IgG2c antibodies in the sera of infected H. saguini-monoassociated mice were significantly increased (P < 0.0001) (Fig. 9). The IgG2c response was significantly higher than the Th2-associated antibody IgG1 response (P < 0.0001), consistent with IgG isotype subclass responses of C57BL IL-10−/− mice to Helicobacter infections (26).
FIG 9.
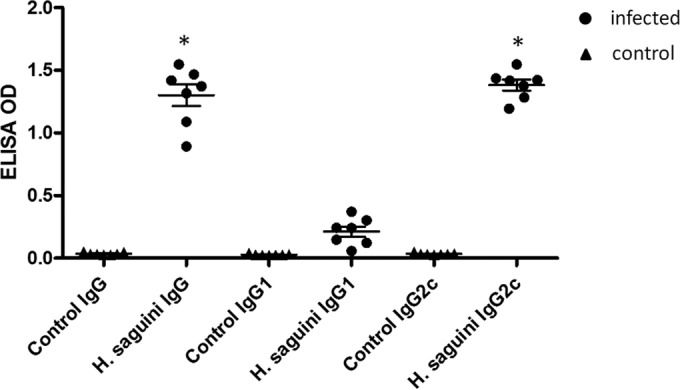
Antibody responses against H. saguini in the sera of GF mice were measured by ELISA. In the H. saguini-monoassociated mice, the total IgG was significantly increased (P < 0.0001); the Th1-associated antibody IgG2c response was significantly higher than the Th2-associated antibody IgG1 response (*, P < 0.0001).
DISCUSSION
To further assess the prevalence of H. saguini infection in CTTs, we surveyed a collection of frozen anal swabs, feces, and colonic biopsy samples previously collected in 2001 and 2006 from a colony in which colitis was endemic. From the 147 samples analyzed from 116 animals, 69% were PCR positive for Helicobacter sp. infection. Representative PCR-positive samples were H. saguini based on 16S rRNA sequence analysis. Given the age of the frozen samples and the difficulty of culturing Helicobacter spp., Helicobacter spp. were successfully cultured only from 11 of the 57 samples; however, in all cases, the cultured Helicobacter sp. was identical to H. saguini by 16S rRNA sequencing. Sequential sampling of selected CTTs indicated that these animals were persistently colonized or, less likely, reinfected. Based on our knowledge of EHS infections in mice and H. macacae infection in rhesus macaques, we believe H. saguini is a persistent infection in CTTs. Whether H. saguini is also present in wild CTTs residing in South America is difficult to assess and remains unknown.
IL-10−/− mice develop spontaneous colitis when housed under conventional or SPF conditions but fail do to so when housed under germfree conditions (32, 33). Thus, without the anti-inflammatory actions of IL-10, an unregulated Th1-mediated immune response against selected enteric bacteria ensues and precipitates colitis characterized histologically by epithelial hyperplasia, mucosal and submucosal inflammation, and ulcerative lesions in the cecal-colonic junction (26, 34–38). EHS infection exacerbates the onset of disease in IL-10−/− mice and thus is a frequently used model to study pathogen-induced IBD and colon cancer. H. hepaticus and H. bilis are the most thoroughly studied EHS; however, other EHS, like H. trogontum, H. typhlonius, H. rodentium, H. mastomyrinus, and H. cinaedi, a human pathogen, can also cause IBD-like disease in immunocompromised mice (9, 11, 34, 38–40). Because EHS-induced colitis in IL-10−/− mice is well characterized, we chose this mouse model to investigate the pathogenic potential of H. saguini.
H. saguini induced typhlocolitis in monoassociated C57BL/129 IL-10−/− mice and elicited pathological lesions consistent with those previously described for EHS-induced colitis in SPF IL-10−/− mice, including cecal and colonic transmural inflammation and epithelial hyperplasia and dysplasia. Interestingly, H. saguini colonized and induced typhlocolitis in monoassociated, but not SPF, mice. This was unexpected considering previous reports suggesting that H. hepaticus infection required other gut microflora in IL-10−/− mice for the induction of colitis. Infection by H. hepaticus produced colitis in conventional or SPF mice, but not in germfree C57BL/129 IL-10−/− mice (41, 42). H. saguini may have failed to colonize SPF mice because of colonization resistance conferred by commensal flora. H. saguini was isolated from a primate and is not a natural colonizer in rodents. In our previous experiments, H. macacae, which was isolated from rhesus monkeys, was inoculated into SPF C57BL/129 IL-10−/− mice and failed to colonize the mice (unpublished data). The different gut flora of primates which allow colonization of Helicobacter spp. in primates, but not mice, could contribute to the inability of specific Helicobacter spp. to colonize mice. Importantly, H. saguini is sufficient by itself to cause IBD in monoassociated IL-10−/− mice, unlike H. hepaticus (41, 42), suggesting that the expression of virulence properties of H. saguini does not require cocolonization of the lower bowel with other flora to initiate an inflammatory response.
The genome of H. saguini with its fusiform morphology is rather large. Nevertheless, other enterohepatic Helicobacter spp. also have draft genome sizes of the same magnitude as that of H. saguini. Helicobacter spp. which have a fusiform morphology tend to have large genomes: H. bilis has a genome size of 2.5 Mb, and H. trogontum has a size of 2.77 Mb. In contrast, curved or spiral-shaped helicobacters have smaller-sized genomes, such as those of H. hepaticus and H. rodentium, which have 1.8-Mb genomes. There are 28 Helicobacter species which have complete or draft genomes available on NCBI. Pseudogene counts range from 16 genes in H. mustelae to 526 genes in H. apodemus. H. saguini has the third-highest pseudogene count at 365 genes, while H. magdeburgensis has the second highest at 398 genes. The average ± standard deviation of pseudogene counts from the 28 genomes is approximately 140 ± 120 genes. So although H. saguini falls into the upper range, having 365 pseudogenes, there is also substantial diversity in pseudogene counts overall within the Helicobacter genomes.
Interestingly, H. saguini is relatively devoid of known EHS virulence factors, particularly cytolethal distending toxin. However, the γ-glutamyl transpeptidase (ggt) gene was detected in the annotated genome of H. saguini. GGT genes from H. pylori, H. suis, H. bilis, and Campylobacter jejuni have been shown to be influential virulence factors required for persistence of the organism in vivo, as well as important in inducing inflammation in vivo (43–46). Also, in vitro data show that GGT inhibits gastrointestinal and lymphocyte cell proliferation as well as having a role in producing reactive free oxygen species (43–46). NF-κB activation and IL-8 secretion in gastrointestinal cell lines may have also been stimulated by GGT expression. Ongoing studies in our laboratory with H. saguini, which persistently colonizes the lower bowel of CTTs, are exploring whether GGT plays a role in H. saguini's pathogenic potential by weakening the intestinal barrier integrity, modulating immune function, and sustaining chronic inflammation.
Chronic intestinal inflammation is well known to increase the risk of colon cancers (10, 47, 48). Chronically inflamed tissues which are continuously regenerating are at an increased risk for mutagenesis and tumor transformation. It has been reported that inflammation-induced cell proliferation greatly potentiates exposure-induced mutations; inflammation can act synergistically with DNA damage to induce mutations that promote cancer and cancer recurrence (49). Reactive oxygen species (ROS) are associated with the inflammatory response and frequently contribute to the tissue-damaging effects of inflammatory reactions. In H. saguini-monoassociated IL-10−/− mice, iNOS expression, a biomarker for nitrosative intestinal damage, also noted in H. hepaticus-induced IBD and colon cancer in Rag2 mice (9, 10, 50), was significantly higher in the IL-10−/− H. saguini-infected mice. Suspected DNA DSBs were detected in the inflamed cecum of monoassociated H. saguini IL-10−/− mice, especially in the regions with high hyperplasia and dysplasia scores. DNA damage in the epithelial cells may lead to mutations and cancer formation in H. saguini-infected mice, and a similar mechanism may be present in the H. saguini-infected CTT colonies with a high incidence of colon cancer.
IBD in CTTs, similar to IBD in humans, is likely multifactorial in etiology but occurs at a higher incidence in CTTs maintained in captivity; CTTs in the wild have a low incidence of mild colitis (2). The precise inflammatory stimuli responsible for IBD in captive CTTs are unknown but may be related to housing conditions, diet, social interaction, and infectious agents. Captivity can also elicit chronic stress, which is known to suppress immune function and increase susceptibility to pathogenic microbes. We have demonstrated that H. saguini persistently colonizes the lower bowel of captive CTTs maintained in a colony, with a high incidence of colitis and colon cancer. H. saguini is also capable of eliciting chronic colitis in monoassociated IL-10−/− mice, has robust in vitro proinflammatory properties, and induces DNA damage. Further, the immune response measured as total IgG and IgG2c isotype was robust and on a scale consistent with data from our prior publication (26). The IgG2c isotype reflects a proinflammatory Th1 response in C57BL/129 IL-10−/− mice, and there is typically an inverse relationship with the Th2-associated IgG1 isotype, which is the reason IgG1 measurements were low.
The role of Helicobacter sp.-induced IBD in humans is unknown, but EHS have been identified in human cases of IBD, as well diarrheal patients (17, 51, 52). Importantly, H. fennelliae and H. cinaedi were originally isolated from the colons of homosexual men with proctitis, who were presumably immunocompromised by HIV infection (15). The H. saguini mouse model described in this report can be used to study the pathogenic potential of EHS isolated from nonhuman primates with IBD. Although new insights into the cause of human IBD were not expressly studied, the isolation of a novel Helicobacter sp. from CTT historically used to study the role of UC and colon cancer serves as a surrogate nonhuman primate model of IBD. Isolation and characterization of EHS in CTT reinforce that there may be a connection between EHS infection and IBD in CTTs and, by extrapolation, also in humans. The phenotypic distinctiveness of H. saguini and the genomic distance between H. saguini and H. jaachi type strains demonstrate that the former is appropriately classified as a novel Helicobacter species.
Description of Helicobacter saguini sp. nov.
Helicobacter saguini (sa′gui.ni. N. L. masc. gen. n. of Saguinus [Oedipus], taxonomic name of cotton-top tamarin from which the bacterium was first isolated). The organism is motile; cells are fusiform with periplasmic fibers, having multiple, bipolar sheathed flagella, and measure 0.5 by 5 μm. The bacterium is Gram negative and nonsporulating; it grows slowly and appears on solid agar as a spreading film on the surface. The bacterium grows at 37o and 42°C, but not at 25°C, under microaerobic conditions, but not aerobically or anaerobically. The bacterium is oxidase, catalase, and γ-glutamyl transpeptidase positive, but urease, indoxyl acetate hydrolysis, and alkaline phosphatase hydrolysis negative. It grows on 1% glycine and is resistant to cephalothin and nalidixic acid. The type strain MIT 97-6194-5 has been deposited in BCCM/LMG Bacteria Collection as LMG 28611. It has a DNA G+C content of 34.6%, and its genome is 2.92 Mb. The 16S rRNA sequence of the type strain has been deposited in GenBank under accession number AF107494.1. The draft genome has been submitted to GenBank under accession number JRMP00000000.
ACKNOWLEDGMENTS
We thank Hans Trüper for providing taxonomic expertise in naming this novel Helicobacter sp., Lenzie Cheaney, Christian Kaufman, and Carlos Umana for technical assistance, and Alyssa Pappa for assistance with manuscript preparation.
REFERENCES
- 1.Johnson LD, Ausman LM, Sehgal PK, King NW Jr. 1996. A prospective study of the epidemiology of colitis and colon cancer in cotton-top tamarins (Saguinus oedipus). Gastroenterology 110:102–115. doi: 10.1053/gast.1996.v110.pm8536845. [DOI] [PubMed] [Google Scholar]
- 2.Wood JD, Peck OC, Tefend KS, Rodriguez MM, Rodriguez MJ, Hernandez CJ, Stonerook MJ, Sharma HM. 1998. Colitis and colon cancer in cotton-top tamarins (Saguinus oedipus oedipus) living wild in their natural habitat. Dig Dis Sci 43:1443–1453. doi: 10.1023/A:1018842210330. [DOI] [PubMed] [Google Scholar]
- 3.Mansfield KG, Lin KC, Xia D, Newman JV, Schauer DB, MacKey J, Lackner AA, Carville A. 2001. Enteropathogenic Escherichia coli and ulcerative colitis in cotton-top tamarins (Saguinus oedipus). J Infect Dis 184:803–807. doi: 10.1086/322990. [DOI] [PubMed] [Google Scholar]
- 4.Johnson LD, Ausman LM, Rolland RM, Chalifoux LV, Russell RG. 2001. Campylobacter-induced enteritis and diarrhea in captive cotton-top tamarins (Saguinus oedipus) during the first year of life. Comp Med 51:257–261. [PubMed] [Google Scholar]
- 5.Saunders KE, Shen Z, Dewhirst FE, Paster BJ, Dangler CA, Fox JG. 1999. Novel intestinal Helicobacter species isolated from cotton-top tamarins (Saguinus oedipus) with chronic colitis. J Clin Microbiol 37:146–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen R, Thomson JM, Fox JG, El-Omar EM, Hold GL. 2011. Could Helicobacter organisms cause inflammatory bowel disease? FEMS Immunol Med Microbiol 61:1–14. doi: 10.1111/j.1574-695X.2010.00744.x. [DOI] [PubMed] [Google Scholar]
- 7.Yu Q, Zhang S, Li L, Xiong L, Chao K, Zhong B, Li Y, Wang H, Chen M. 2015. Enterohepatic Helicobacter species as a potential causative factor in inflammatory bowel disease: a meta-analysis. Medicine 94:e1773. doi: 10.1097/MD.0000000000001773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erdman SE, Poutahidis T, Tomczak M, Rogers AB, Cormier K, Plank B, Horwitz BH, Fox JG. 2003. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am J Pathol 162:691–702. doi: 10.1016/S0002-9440(10)63863-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Erdman SE, Rao VP, Poutahidis T, Rogers AB, Taylor CL, Jackson EA, Ge Z, Lee CW, Schauer DB, Wogan GN, Tannenbaum SR, Fox JG. 2009. Nitric oxide and TNF-alpha trigger colonic inflammation and carcinogenesis in Helicobacter hepaticus-infected, Rag2-deficient mice. Proc Natl Acad Sci U S A 106:1027–1032. doi: 10.1073/pnas.0812347106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mangerich A, Knutson CG, Parry NM, Muthupalani S, Ye W, Prestwich E, Cui L, McFaline JL, Mobley M, Ge Z, Taghizadeh K, Wishnok JS, Wogan GN, Fox JG, Tannenbaum SR, Dedon PC. 2012. Infection-induced colitis in mice causes dynamic and tissue-specific changes in stress response and DNA damage leading to colon cancer. Proc Natl Acad Sci U S A 109:E1820–E1829. doi: 10.1073/pnas.1207829109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nguyen DD, Muthupalani S, Goettel JA, Eston MA, Mobley M, Taylor NS, McCabe A, Marin R, Snapper SB, Fox JG. 2013. Colitis and colon cancer in WASP-deficient mice require helicobacter species. Inflamm Bowel Dis 19:2041–2050. doi: 10.1097/MIB.0b013e318295fd8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lertpiriyapong K, Handt L, Feng Y, Mitchell TW, Lodge KE, Shen Z, Dewhirst FE, Muthupalani S, Fox JG. 2014. Pathogenic properties of enterohepatic Helicobacter spp. isolated from rhesus macaques with intestinal adenocarcinoma. J Med Microbiol 63:1004–1016. doi: 10.1099/jmm.0.072462-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marini RP, Muthupalani S, Shen Z, Buckley EM, Alvarado C, Taylor NS, Dewhirst FE, Whary MT, Patterson MM, Fox JG. 2010. Persistent infection of rhesus monkeys with “Helicobacter macacae” and its isolation from an animal with intestinal adenocarcinoma. J Med Microbiol 59:961–969. doi: 10.1099/jmm.0.019117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fox JG, Boutin SR, Handt LK, Taylor NS, Xu S, Rickman B, Marini RP, Dewhirst FE, Paster BJ, Motzel S, Klein HJ. 2007. Isolation and characterization of a novel helicobacter species, “Helicobacter macacae,” from rhesus monkeys with and without chronic idiopathic colitis. J Clin Microbiol 45:4061–4063. doi: 10.1128/JCM.01100-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Totten PA, Fennell CL, Tenover FC, Wezenberg JM, Perine PL, Stamm WE, Holmes KK. 1985. Campylobacter cinaedi (sp. nov.) and Campylobacter fennelliae (sp. nov.): two new Campylobacter species associated with enteric disease in homosexual men. J Infect Dis 151:131–139. doi: 10.1093/infdis/151.1.131. [DOI] [PubMed] [Google Scholar]
- 16.Flores BM, Fennell CL, Kuller L, Bronsdon MA, Morton WR, Stamm WE. 1990. Experimental infection of pig-tailed macaques (Macaca nemestrina) with Campylobacter cinaedi and Campylobacter fennelliae. Infect Immun 58:3947–3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thomson JM, Hansen R, Berry SH, Hope ME, Murray GI, Mukhopadhya I, McLean MH, Shen Z, Fox JG, El-Omar E, Hold GL. 2011. Enterohepatic helicobacter in ulcerative colitis: potential pathogenic entities? PLoS One 6:e17184. doi: 10.1371/journal.pone.0017184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fox JG, Dewhirst FE, Shen Z, Feng Y, Taylor NS, Paster BJ, Ericson RL, Lau CN, Correa P, Araya JC, Roa I. 1998. Hepatic Helicobacter species identified in bile and gallbladder tissue from Chileans with chronic cholecystitis. Gastroenterology 114:755–763. doi: 10.1016/S0016-5085(98)70589-X. [DOI] [PubMed] [Google Scholar]
- 19.Shen Z, Xu S, Dewhirst FE, Paster BJ, Pena JA, Modlin IM, Kidd M, Fox JG. 2005. A novel enterohepatic Helicobacter species “Helicobacter mastomyrinus” isolated from the liver and intestine of rodents. Helicobacter 10:59–70. doi: 10.1111/j.1523-5378.2005.00292.x. [DOI] [PubMed] [Google Scholar]
- 20.Kaur T, Singh J, Huffman MA, Petrzelkova KJ, Taylor NS, Xu S, Dewhirst FE, Paster BJ, Debruyne L, Vandamme P, Fox JG. 2011. Campylobacter troglodytis sp. nov., isolated from feces of human-habituated wild chimpanzees (Pan troglodytes schweinfurthii) in Tanzania. Appl Environ Microbiol 77:2366–2373. doi: 10.1128/AEM.01840-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shen Z, Feng Y, Sheh A, Everitt J, Bertram F, Paster BJ, Fox JG. 2015. Isolation and characterization of a novel Helicobacter species, Helicobacter jaachi sp. nov., from common marmosets (Callithrix jaachus). J Med Microbiol 64:1063–1073. doi: 10.1099/jmm.0.000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sheh A, Piazuelo MB, Wilson KT, Correa P, Fox JG. 2013. Draft genome sequences of Helicobacter pylori strains isolated from regions of low and high gastric cancer risk in Colombia. Genome Announc 1:e00736-13. doi: 10.1128/genomeA.00736-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zerbino DR, Birney E. 2008. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O'Neill K, Resch W, Resenchuk S, Schafer S, Tolstoy I, Tatusova T. 2009. The National Center for Biotechnology Information's Protein Clusters Database. Nucleic Acids Res 37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Meier-Kolthoff JP, Auch AF, Klenk HP, Goker M. 2013. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shen Z, Feng Y, Rogers AB, Rickman B, Whary MT, Xu S, Clapp KM, Boutin SR, Fox JG. 2009. Cytolethal distending toxin promotes Helicobacter cinaedi-associated typhlocolitis in interleukin-10-deficient mice. Infect Immun 77:2508–2516. doi: 10.1128/IAI.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan V, Crocetti G, Grehan M, Zhang L, Danon S, Lee A, Mitchell H. 2005. Visualization of Helicobacter species within the murine cecal mucosa using specific fluorescence in situ hybridization. Helicobacter 10:114–124. doi: 10.1111/j.1523-5378.2005.00298.x. [DOI] [PubMed] [Google Scholar]
- 28.Meier-Kolthoff JP, Klenk HP, Goker M. 2014. Taxonomic use of DNA G+C content and DNA-DNA hybridization in the genomic age. Int J Syst Evol Microbiol 64:352–356. doi: 10.1099/ijs.0.056994-0. [DOI] [PubMed] [Google Scholar]
- 29.Ivashkevich A, Redon CE, Nakamura AJ, Martin RF, Martin OA. 2012. Use of the gamma-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Lett 327:123–133. doi: 10.1016/j.canlet.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Turinetto V, Giachino C. 2015. Multiple facets of histone variant H2AX: a DNA double-strand-break marker with several biological functions. Nucleic Acids Res 43:2489–2498. doi: 10.1093/nar/gkv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koeppel M, Garcia-Alcalde F, Glowinski F, Schlaermann P, Meyer TF. 2015. Helicobacter pylori infection causes characteristic DNA damage patterns in human cells. Cell Rep 11:1703–1713. doi: 10.1016/j.celrep.2015.05.030. [DOI] [PubMed] [Google Scholar]
- 32.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. 1993. Interleukin-10-deficient mice develop chronic enterocolitis. Cell 75:263–274. doi: 10.1016/0092-8674(93)80068-P. [DOI] [PubMed] [Google Scholar]
- 33.Sellon RK, Tonkonogy S, Schultz M, Dieleman LA, Grenther W, Balish E, Rennick DM, Sartor RB. 1998. Resident enteric bacteria are necessary for development of spontaneous colitis and immune system activation in interleukin-10-deficient mice. Infect Immun 66:5224–5231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kullberg MC, Jankovic D, Gorelick PL, Caspar P, Letterio JJ, Cheever AW, Sher A. 2002. Bacteria-triggered CD4+ T regulatory cells suppress Helicobacter hepaticus-induced colitis. J Exp Med 196:505–515. doi: 10.1084/jem.20020556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kullberg MC, Jankovic D, Feng CG, Hue S, Gorelick PL, McKenzie BS, Cua DJ, Powrie F, Cheever AW, Maloy KJ, Sher A. 2006. IL-23 plays a key role in Helicobacter hepaticus-induced T cell-dependent colitis. J Exp Med 203:2485–2494. doi: 10.1084/jem.20061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ray A, Basu S, Gharaibeh RZ, Cook LC, Kumar R, Lefkowitz EJ, Walker CR, Morrow CD, Franklin CL, Geiger TL, Salzman NH, Fodor A, Dittel BN. 2015. Gut microbial dysbiosis due to Helicobacter drives an increase in marginal zone B cells in the absence of IL-10 signaling in macrophages. J Immunol 195:3071–3085. doi: 10.4049/jimmunol.1500153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chichlowski M, Westwood GS, Abraham SN, Hale LP. 2010. Role of mast cells in inflammatory bowel disease and inflammation-associated colorectal neoplasia in IL-10-deficient mice. PLoS One 5:e12220. doi: 10.1371/journal.pone.0012220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fox JG, Gorelick PL, Kullberg MC, Ge Z, Dewhirst FE, Ward JM. 1999. A novel urease-negative Helicobacter species associated with colitis and typhlitis in IL-10-deficient mice. Infect Immun 67:1757–1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shen Z, Feng Y, Rickman B, Fox JG. 2015. Helicobacter cinaedi induced typhlocolitis in Rag-2-deficient mice. Helicobacter 20:146–155. doi: 10.1111/hel.12179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eaton KA, Opp JS, Gray BM, Bergin IL, Young VB. 2011. Ulcerative typhlocolitis associated with Helicobacter mastomyrinus in telomerase-deficient mice. Vet Pathol 48:713–725. doi: 10.1177/0300985810383876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dieleman LA, Arends A, Tonkonogy SL, Goerres MS, Craft DW, Grenther W, Sellon RK, Balish E, Sartor RB. 2000. Helicobacter hepaticus does not induce or potentiate colitis in interleukin-10-deficient mice. Infect Immun 68:5107–5113. doi: 10.1128/IAI.68.9.5107-5113.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whary MT, Taylor NS, Feng Y, Ge Z, Muthupalani S, Versalovic J, Fox JG. 2011. Lactobacillus reuteri promotes Helicobacter hepaticus-associated typhlocolitis in gnotobiotic B6.129P2-IL-10tm1Cgn (IL10−/−) mice. Immunology 133:165–178. doi: 10.1111/j.1365-2567.2011.03423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Floch P, Pey V, Castroviejo M, Dupuy JW, Bonneu M, de la Guardia AH, Pitard V, Megraud F, Lehours P. 2014. Role of Campylobacter jejuni gamma-glutamyl transpeptidase on epithelial cell apoptosis and lymphocyte proliferation. Gut Pathog 6:20. doi: 10.1186/1757-4749-6-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Javed S, Mejias-Luque R, Kalali B, Bolz C, Gerhard M. 2013. Helicobacter bilis gamma-glutamyltranspeptidase enhances inflammatory stress response via oxidative stress in colon epithelial cells. PLoS One 8:e73160. doi: 10.1371/journal.pone.0073160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rossi M, Bolz C, Revez J, Javed S, El-Najjar N, Anderl F, Hyytiainen H, Vuorela P, Gerhard M, Hanninen ML. 2012. Evidence for conserved function of gamma-glutamyltranspeptidase in Helicobacter genus. PLoS One 7:e30543. doi: 10.1371/journal.pone.0030543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang G, Ducatelle R, De Bruyne E, Joosten M, Bosschem I, Smet A, Haesebrouck F, Flahou B. 2015. Role of gamma-glutamyltranspeptidase in the pathogenesis of Helicobacter suis and Helicobacter pylori infections. Vet Res 46:31. doi: 10.1186/s13567-015-0163-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grivennikov SI. 2013. Inflammation and colorectal cancer: colitis-associated neoplasia. Semin Immunopathol 35:229–244. doi: 10.1007/s00281-012-0352-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Francescone R, Hou V, Grivennikov SI. 2015. Cytokines, IBD, and colitis-associated cancer. Inflamm Bowel Dis 21:409–418. doi: 10.1097/MIB.0000000000000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kiraly O, Gong G, Olipitz W, Muthupalani S, Engelward BP. 2015. Inflammation-induced cell proliferation potentiates DNA damage-induced mutations in vivo. PLoS Genet 11:e1004901. doi: 10.1371/journal.pgen.1004901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Knutson CG, Mangerich A, Zeng Y, Raczynski AR, Liberman RG, Kang P, Ye W, Prestwich EG, Lu K, Wishnok JS, Korzenik JR, Wogan GN, Fox JG, Dedon PC, Tannenbaum SR. 2013. Chemical and cytokine features of innate immunity characterize serum and tissue profiles in inflammatory bowel disease. Proc Natl Acad Sci U S A 110:E2332–E2341. doi: 10.1073/pnas.1222669110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Fox JG. 2002. The non-H. pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut 50:273–283. doi: 10.1136/gut.50.2.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Castano-Rodriguez N, Kaakoush NO, Lee WS, Mitchell HM. 27 October 2015. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut doi: 10.1136/gutjnl-2015-310545. [DOI] [PubMed] [Google Scholar]