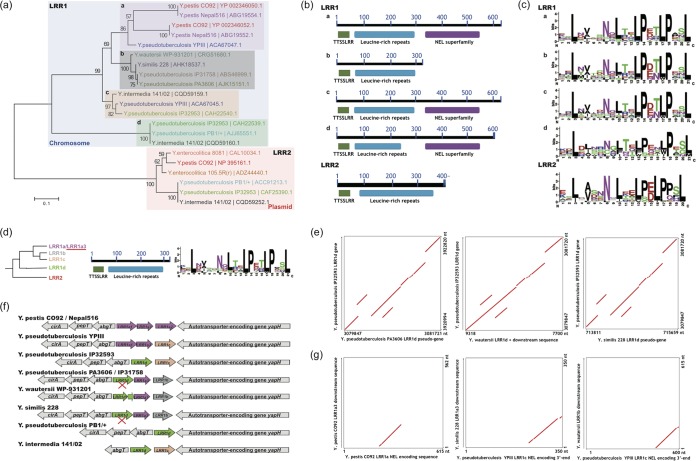
FIG 1.
Screening and sequence analysis of Yersinia LRR proteins. (a) Distribution and phylogenetic analysis of Yersinia LRR proteins. Proteins from the same strain are highlighted in the same color. Sequences with different background colors represent different subgroups. The neighbor-joining distance was calculated and bootstrapping tests performed with MEGA6.0 (see Materials and Methods), and the bootstrapping consistence percentage is indicated for each node. (b) LRR protein domain analysis. Conserved domains and their within-protein locations are shown. (c) Repeat-unit consensus motifs of Yersinia LRR proteins. (d) LRR1a3 sequence features. (Left) Phylogenetic diagram; (middle) domain composition diagram; (right) repeat-unit consensus motif. (e) Comparison of LRR1d loci between Yersinia strains with an LRR1d gene or pseudogene. The collinear relationship is shown with red lines or plots. (f) Synteny analysis of LRR loci of different Yersinia strains. Pseudogenes are marked with “X's.” LRR genes of different subgroups are highlighted in different colors. (g) Nucleotide-level NEL domain relics identified from genome comparisons. The collinear relationship between compared sequences is shown in red.