Figure 4.
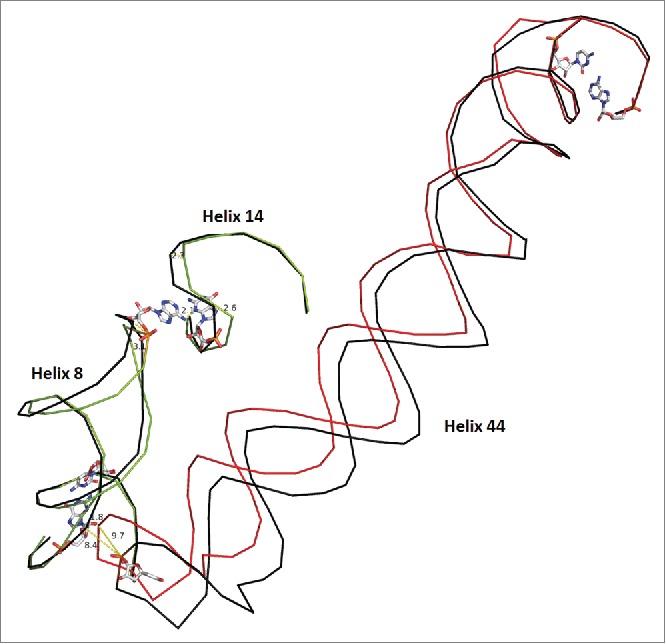
Bridge B8 in the SSU. Helix 44 moves by ∼10 Angstroms in the EF-Tu bound ribosome vs. the unbound molecule. The unbound state is shown in black for each helix with the bound state in a unique color for each protein. Pivoting bases are shown as stick models. Helix h44 contacts h8 and induces a 3.0 Angstrom change in the final residue of h8 which moves 2.1 Angstroms closer to h14. Although h14 is a relatively small helix, it exhibits some conformational freedom, as a result. Helix 10, in proximity to the pivots, displays minor motion and connects h17 to bridge B8 as shown in supplemental Figs. S5 and S6. Overall, the scheme shows a connected network of interactions, which explain partially, the structural consequence of EF-Tu ribosome association. Structure 2J00 is in black and blue. Structure 4ABR is in red and green)
