Figure 5.
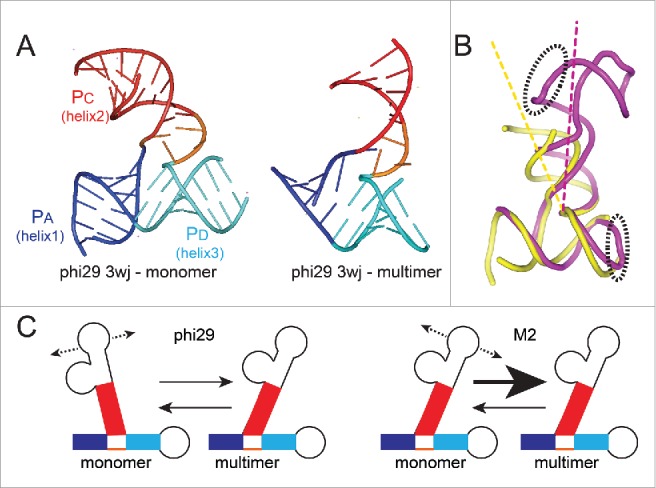
Conformation of 3wjs and putative relationship to multimer formation. (A) Left: Ribbon diagram of the structure of the isolated 3wj from phi29 pRNA, solved by X-ray crystallography (PDB: 4KZ2).54 This RNA did not have the kissing-loop interaction elements and thus was unable to self-associate and is considered to be the structure in monomer form. Right: Ribbon diagram of the structure of the 3wj from phi29 pRNA, within the context of an RNA that was engineered for crystallization but could still self-associate, forming a tetramer in the crystal (PDB: 3R4F).14 For the latter structure, only the 3wj is shown. For both structures, the 3 helices that emerge from the junction are shown in blue, red, and cyan and labeled. (B) Superimposed structures of the 3wj in monomer (yellow) and multimer (magenta) form, using the structures from panel (A). The entire structure is shown, and the elements that form the intermolecular interaction important for self-association are in dashed ellipses. (C) Model for how the 3wj conformation may affect multimer formation. The 3wjs differ in their conformations. If they are conformationally dynamic, we propose the conformational equilibria of sampled states differs. For phi29 pRNA, this conformational equilibrium may be more different from the multimer state than it is in the M2 pRNA.
