Figure 3.
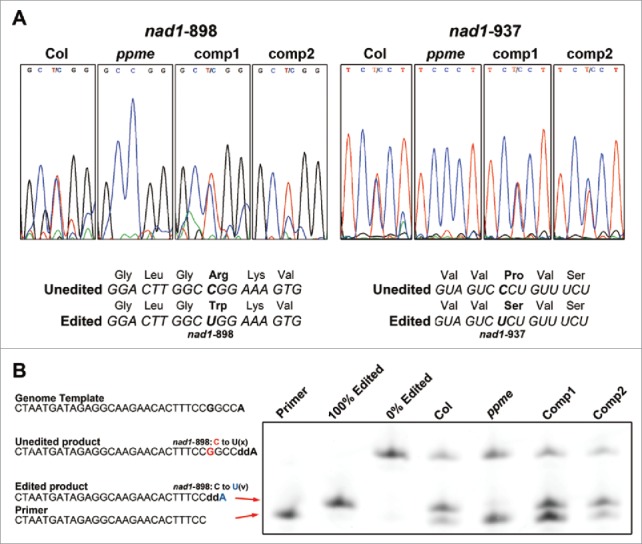
Defective editing at mitochondrial nad1-898 and nad1-937 RNA editing sites in homozygous ppme−/− seedlings. (A) Sequencing of cDNAs from 14-day-old wild-type, homozygous ppme−/−, and complemented seedlings for assessment of nad1-898 and nad1-937 RNA editing efficiencies in ppme−/− and complementation lines. The middle cytosine or thymine in each panel is the position of the nad1-898 or nad1-937 site, respectively. The lower panel depicts the predicted amino acid changes after nad1-898 and nad1-937 editing. In wild-type mitochondria, nad1-898 and nad1-937 RNA editing causes amino acid changes from Arg to Trp and from Pro to Ser, respectively, in the NAD1 protein after translation. The bold cytosine (C) and uracil (U) indicate the nad1-898 and nad1-937 editing sites, respectively. (B) Poisoned primer extension assay of the nad1-898 editing site. The edited products were terminated earlier than the unedited products by stopping the reaction with ddATP. The edited and unedited products were separated in a sequencing gel and visualized by detection of FAM fluorescence signals. The edited products are from wild-type and ppme−/− seedlings and seedlings from 2 complementation lines.
