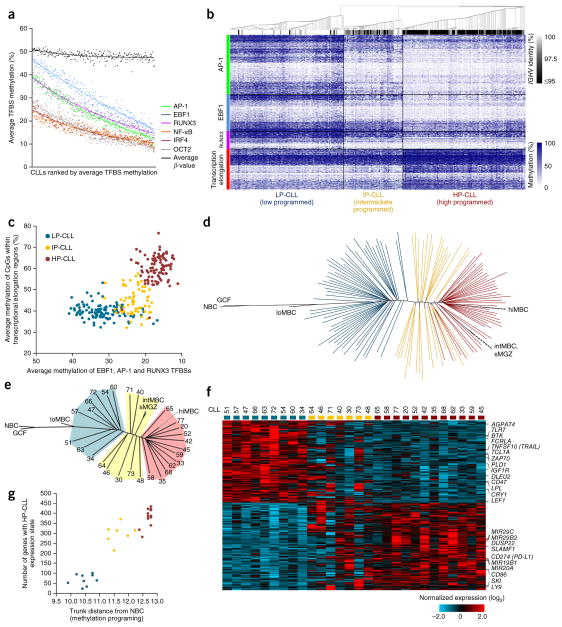
Figure 2.
Evaluation of the maturation state of CLL using DNA methylation. (a) The average level of methylation at transcription factor binding sites for all six significantly programmed transcription factors in 267 CLL samples assessed by 450K array. Samples are ranked according to the average methylation level for all transcription factor binding sites. The average β value (global methylation) is also shown for each sample. (b) Heat map showing the methylation at all high-confidence AP-1, EBF1 and RUNX3 binding sites and in hypermethylated transcriptional elongation regions in 267 CLL samples. Consensus clustering of CLL samples identifies three methylation subtypes (LP-CLL, IP-CLL and HP-CLL). The level of IGHV homology (identity) for each CLL is indicated. (c) Scatterplot displaying the average methylation of AP-1, EBF1 and RUNX3 binding sites versus hypermethylated transcriptional elongation regions. (d) DNA methylation phylogenetic tree diagram of the DKFZ CLL sample cohort (n = 128) and normal B cell subtypes generated using AP-1, EBF1 and RUNX3 binding sites and hypermethylated transcriptional elongation regions. Each line represents a CLL sample; CpG methylation values for normal samples were averaged according to subtype. Colors indicate CLL subtypes as in c. (e) DNA methylation phylogenetic tree diagram of the CLL samples selected for RNA-seq analysis. Patient numbers are indicated for each sample; shaded areas represent the CLL subtypes. (f) Heat map of 459 genes differentially expressed in the LP-CLL and HP-CLL subtypes (FDR q < 0.05). Samples are ordered according to their position within the phylogenetic analysis in e. Genes that have been previously reported to be differentially expressed in the IGHV subtypes or to have a role in CLL pathogenesis are also indicated. (g) Scatterplot showing the correlation between the degree of methylation maturation (as assessed by the trunk distance from naive B cells in the phylogenetic analysis) and the acquisition of the HP-CLL expression state.