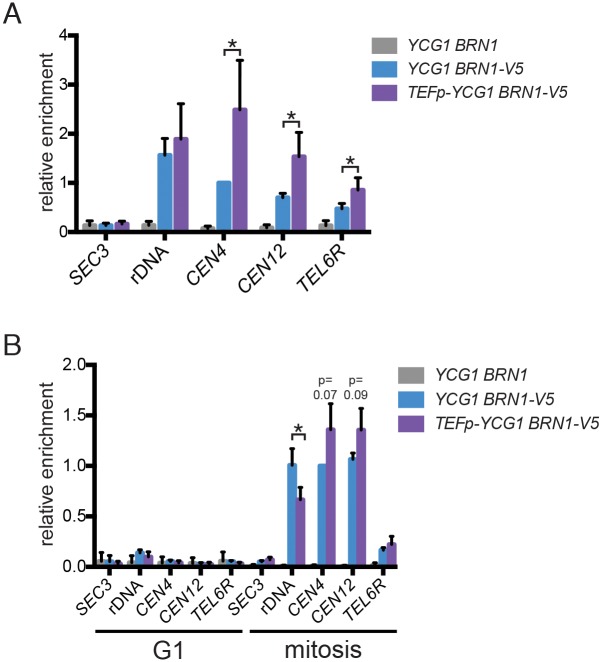
Fig 8. Increasing Ycg1 expression in G1 does not promote condensin recruitment to mitosis-enriched sites.
(A) ChIP-qPCR of Brn1-3V5 from asynchronous YCG1 (YTD297) and TEFp-YCG1 (YTD342) strains, as well a wild-type strain lacking a 3V5 tag (YTD33) as a negative control. Brn1 enrichment at the rDNA (replication fork barrier), the centromeres of chromosome IV and XII (CEN4 and CEN12), the subtelomeric region on the right arm of chromosome VI (TEL6R), and a condensin-depleted region in the SEC3 gene on chromosome V (SEC3) was quantified by qPCR. Data was normalized to Brn1 enrichment at the CEN4 locus in YCG1 BRN1-3V5 cells in each experiment and represent mean values of 4 biological replicates +/- 1 standard deviation. Significance was determined by an unpaired t-test (*p<0.05). (B) ChIP-qPCR as in (A), except that strains were arrested in G1 with alpha-factor or in metaphase with nocodazole, prior to processing. Data was normalized to Brn1 enrichment at the CEN4 locus in YCG1 BRN1-3V5 cells arrested in mitosis in each experiment, and represent mean values of 3 biological replicates +/- 1 standard deviation. Significance was determined by an unpaired t-test (*p<0.05). For centromeric loci that did not show statistically significant increases in binding, the p-values are indicated. Flow cytometry plots confirming cell-cycle positions for all experiments are shown in S7 Fig.